Introduction: In the last decades, the development of controlled drug delivery systems has attracted considerable attention in biochemical and therapeutic treatments. The synthesis of hydrogels as devices functionalized through cleavable linkers gives the opportunity to produce vehicles capable to tune drug release in targeted conditions, also maintaining the drug level within a desired range for a sustained period of time. However, respect to the good results obtained with antibodies and peptides[1], several critical aspects are related to the fact that drug release from 3D matrices is mostly driven by very quickly pure diffusion mechanism[2]. In this work, we propose the microwave-assisted synthesis of hydrogels functionalized with pH-sensitive linkers, ester and hydrazone, and the study of multiple tunable delivery of rhodamine B (RhB) conjugated with the functionalized polymeric network[3],[4]. RhB choice was due to its resemblance to many hydrophilic analgesic and anti-inflammatory drugs[5].
Materials and Methods: Polymeric functionalization occurred on polyethylene glycol (PEG) chains. To synthetize the final hydrogels with ester cleavable linkers (ACPEG-e hydrogels), PEG reacted with RhB acid chloride through esterification reaction; whereas hydrogels with hydrazone functionality (ACPEG-h hydrogels) were obtained using PEG modified aldehyde reacted with RhB hydrazine derivative. Hydrogel synthesis occurred via polycondensation reaction between the carboxyl groups of carbomer 974P and hydroxyl groups of PEG, PEG grafting RhB and agarose.
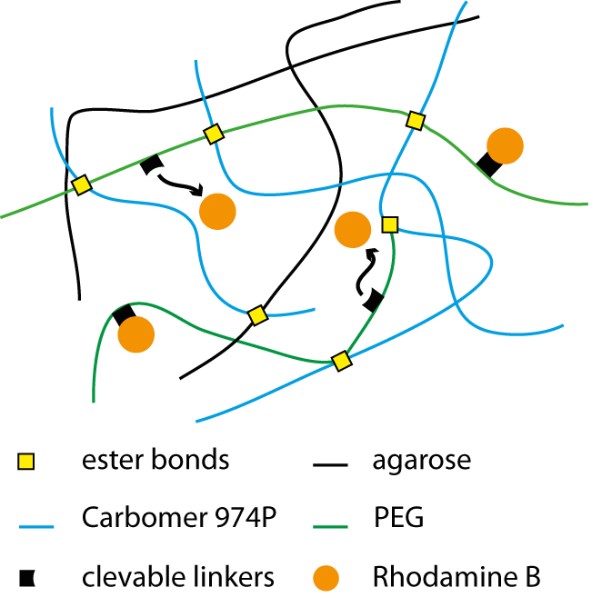
Figure 1. Representation of hydrogels modified with rhodamine-binding cleavable linker. Transient association between rhodamine and PEG modulates release of the drug mimetic from the matrix.
Release experiments of RhB by hydrogels were performed at 37°C at pH 7.4 and 8.5; the released amount was measured by UV spectroscopy at 570 nm and determined referring to the standard calibration curve for RhB.
Results and Discussion: 1H-NMR and FT-IR analyses confirmed the synthesis of the cleavable linkers modified hydrogels. The percentage of released RhB is defined as the ratio between the released amount and the total amount linked to the polymeric scaffold. The comparison of the release profiles associated to ester and hydrazone functionalization and the sample with RhB only physically entrapped within 3D network (ACPEG hydrogels) shows that drug release is prolonged for a much longer period in the functionalized hydrogels. A linear plot of these release profiles at both pH illustrates a double diffusion regime with different slopes. The transition and duration of the two regimes depends on the nature of the functionalization. These regimes probably correspond to: (i) ester/hydrazone bonds present at the interface hydrogel/water that could be easily cleavable and (ii) ester/hydrazone bonds present in the inner core.
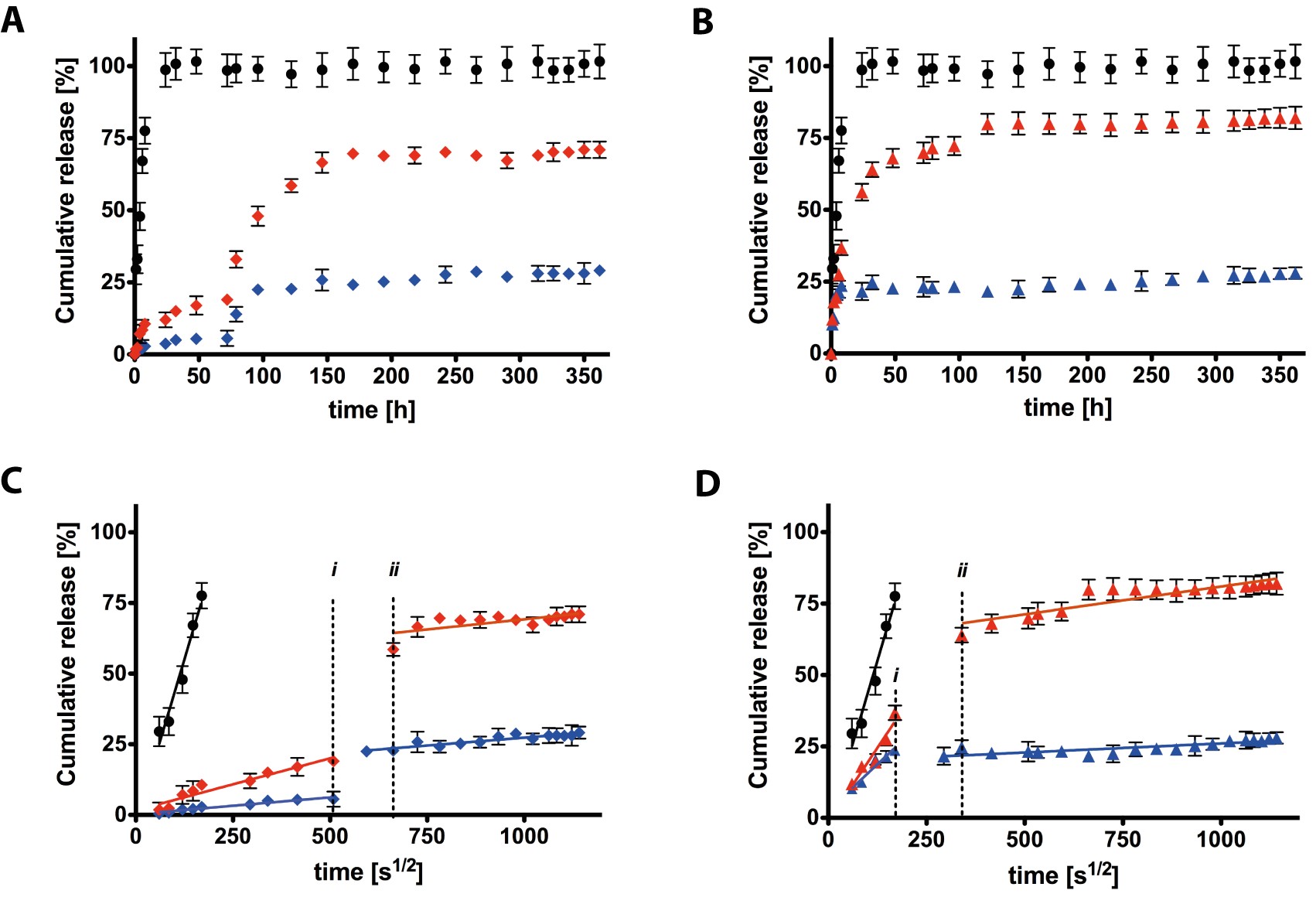
Figure 2. (A) In vitro release profile of rhodamine delivered from ACPEG (circle), ACPEG-e at pH 7.4 (diamond, blue) and ACPEG-e at pH 8.5 (diamond, red) hydrogels. (B) In vitro release profile of rhodamine delivered from ACPEG (circle), ACPEG-h at pH 7.4 (triangle, blue) and ACPEG-h at pH 8.5 (triangle, red) hydrogels. (C) The slope of the rhodamine release from ACPEG (circle), ACPEG-e at pH 7.4 (diamond, blue) and ACPEG-e at pH 8.5 (diamond, red) hydrogels against the square root time. (D) The slope of the rhodamine release from ACPEG (circle), ACPEG-h at pH 7.4 (triangle, blue) and ACPEG-h at pH 8.5 (triangle, red) hydrogels against the square root time is representative of the Fickian diffusion coefficient of NPs in gels (p < 0.0001 between all of the groups).
Conclusion: Microwave-assisted hydrogels composed by PEG modified with ester or hydrazone groups grafting RhB allow to tune the release of the drug load upon different physiological pH conditions. Moreover our method of synthesis yields sterile polymeric networks in absence of organic solvent and byproducts.
References:
[1] A. Sacchetti, E. Mauri, M. Sani, M. Masi and F. Rossi, Tetrahedron Lett., 2014, 55, 6817-6820
[2] F. Rossi, F. Castiglione, M. Ferro, P. Marchini, E. Mauri, M. Moioli, A. Mele, M. Masi, ChemphysChem 2015, 16(13), 2818
[3] D. Ossipov, S. Kootala, Z. Y. Yi, X. Yang and J. Hilborn, Macromolecules, 2013, 46, 4105-4113
[4] W. Khan, S. Farah, A. Nyska and A. J. Domb, J. Control. Release, 2013, 168, 70-76
[5] C. L. Modery-Pawlowski, A. M. Master, V. Pan, G. P. Howard and A. Sen Gupta, Biomacromolecules, 2013, 14, 910-919