1
Department of Bioelectronics, FKE, Vienna University of Technology, Vienna, Austria
2
Bioelectronics, Center for Brain Research, Medical University of Vienna, Vienna, Austria
3
Department of Neurobiology, University of Oldenburg, Oldenburg, Germany
4
Max-Planck-Institute of Biochemistry, Muscle Dynamics, Martinsried, Germany
Genetic mutants are invaluable for understanding the development, physiology and behaviour of Drosophila. Modern molecular genetic techniques enable the rapid generation of large numbers of different mutants. To phenotype these mutants sophisticated microscopy techniques are required, ideally allowing the 3D-reconstruction of the anatomy of an adult fly from a single scan. Ultramicroscopy enables up to cm fields of view, whilst providing micron resolution. In this paper, we present ultramicroscopy reconstructions of the flight musculature, the nervous system, and the digestive tract of entire, chemically cleared, drosophila in autofluorescent light. The 3D-reconstructions thus obtained verify that the anatomy of a whole fly, including the filigree spatial organization of the direct flight muscles, can be analysed from a single ultramicroscopy reconstruction. The recording procedure, including 3D-reconstruction using standard software, takes no longer than 30 min. Additionally, image segmentation, which would allow for further quantitative analysis, was performed.
For several decades, Drosophila melanogaster serves as an outstanding model organism in genetic research. Its high fecundity and its simple cultivation make Drosophila optimally suited for high-throughput screening of the many defined genetic aberrations generated by modern molecular biology approaches. Ideally, a microscopy device for high-throughput phenotyping of Drosophila mutants should allow the 3D-reconstruction of virtually the whole anatomy of a fly in a single scan taking no longer than several minutes. Standard confocal or two photon microscopes provide the required resolution, but since both work best with high magnification objectives the provided fields of view are too small to image a complete fly in a single pass. Ultramicroscopy (Figure 1
) works also well with low power objectives, providing excellent spatial resolution and optical sectioning quality, comparable to confocal microscopy (Dodt et al., 2007
; Jährling et al., 2008
). In this paper we present 3D-reconstructions and segmentations of Drosophila organs obtained from an ultramicroscopy scan of an intact, chemically cleared adult fly.
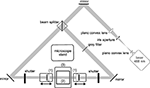
Figure 1. Standard configuration of an ultramicroscopy setup. A laser beam is expanded and homogenized by two convex lenses and then split into two separate light pathways. Both beams are focussed by cylindrical lenses and slit apertures (1), forming a thin light sheet. The light sheet illuminates the specimen placed in a transparent specimen chamber (2) filled with clearing solution. Hence, fluorescence is only exhibited in those parts of the specimen, which are in the focal plane. The fluorescence image is projected to a camera target using a microscope objective, and the excitation light is blocked by a matched optical band pass filter. By stepping the specimen chamber vertically through the light sheet using a computer controlled jack (3), a stack of optical slices is generated.
A detailed anatomical atlas about the internal anatomy of Drosophila was first presented by Miller (1950)
, and later extended by Hartenstein (1993)
. The basic organ systems of Drosophila are the nervous system, the intestinal tract and the musculature. The Drosophila’s muscular system consists of multiple contractile fibres arranged in distinct groups or layers (Miller, 1950
). In the thorax flight, jump and leg muscles are the most prominent. Flight muscles are classified into direct flight muscles (DFMs) and indirect flight muscles (IFMs), according to their functional role in wing movement. The large IFMs of the mesothorax function as a single contractile unit, generating the main propulsion during flight (Dutta et al., 2004
). They are formed by the dorsal longitudinal muscles (DLMs) consisting of six fibres, and three groups of dorsal–ventral muscles (DVM-I, DVM-II, and DVM-III) in each half of the thorax. DVM-I consists of three fibres, DVM-II and DVM-III consist of two fibres, each (Dutta et al., 2004
). The largest muscle of the mesothorax is the tergal depressor of the trochanter (TDT), also called jump muscle enabling jump start of the fly (Dutta et al., 2004
). Unlike the IFMs, which generate the power for the wing beat, the DFMs are responsible for adjusting the orientation of the wings during flight. They are attached to the wing base in such a way that they can generate subtle torsions of the wings, which are responsible for controlling the direction of flight. Due to their filigree structure and their complex spatial arrangement, 3D-reconstructions of the DFMs in the entire fly are challenging, demonstrating the strengths of ultramicroscopy. The central nervous system (CNS) of Drosophila comprises the dorsally located brain enclosed by the head capsule, the thoracico-abdominal ganglion (ThAGl), and the cervical connective (CN), which connects the brain with the ThAGl (Hartenstein, 1993
). In Drosophila the different thoracic ganglia are merged into two bonded masses of neural tissue, no ganglia exist in the abdomen (Miller, 1950
).
3D-reconstructions of chemically cleared entire Drosophila were performed, using the ultramicroscopy setup described in Becker et al. (2008)
. The obtained reconstructions give detailed insight into the anatomy of the flight musculature, the nervous system, and the intestinal tract.
Preparation and Clearing
Adult white eyed Drosophila, w[1118], were killed by ether and fixed at 4°C in 4% paraformaldehyde overnight. Flies were dehydrated in a graded ethanol series (50, 70, 96, 100% for 1 h, last step overnight), and incubated in clearing solution, consisting of two parts benzyl benzoate and one part benzyl alcohol (BABB, Spalteholz, 1914
) for at least 4 h, until they became almost transparent (Figure 2
).
Figure 2. White eyed Drosophila, placed on a Siemens star after dehydration and clearing. The fly is nearly completely translucent. Length of scale bar 1 mm.
Ultramicroscopy
For ultramicroscopy, the setup described in Becker et al. (2008)
was used. Imaging was performed by exciting autofluorescence using a 488-nm, 200 mW diode laser (Sapphire, Coherent, Germany). Images were recorded using a 10× objective (N.A. 0.3), and a CoolSnap K4 camera with 2048 × 2048 pixels (Roper Scientific, Germany).
Segmentation and 3D-Reconstruction
Manual image segmentation was performed, based on the visual shape of anatomical structures of interest. These structures were marked using an interactive pen display (Wacom Cintiq 12WX, Germany) in three orthogonal spatial orientations. The borders of the encircled structures were smoothed using a 6 × 6 Gaussian filter. The visualization software Amira 5.2 (Visage Imaging, Germany), running on a computer with two quad-core processors a 2.5-GHz, 32 GB RAM, and an FX-5800 (NVIDIA, Germany) graphic processor board, was used for all image processing.
An adult fly was three dimensionally reconstructed from an ultramicroscopy stack consisting of 579 images of 2048 × 2048 pixels each, and 1.66 μm vertical spacing (Figure 3
). The surface (Figure 3
A), the situs of various inner organs, and the musculature (Figures 3
B,D) of the entire fly is demonstrated. Figures 3
B,C show the six pairs of DLMs forming the IFMs. The brain and the ThAGl are well visible. The six DFMs attached to the base of each wing (Ghazi et al., 2000
) are easily addressed as DFM49–DFM54 (D). DFM52 can only rudimentarily be identified, because it is clipped by the viewing plane.
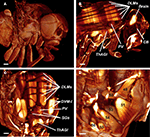
Figure 3. (A) Reconstructed surface of an entire fly. Scale bar 100 μm. (B) Sagittal view of the fly’s inner anatomy, showing parts of the flight muscles, the nervous, and the cardiac system. DLMs, dorso longitudinal muscles; ThAGl, thoracico-abdominal ganglion; PV, proventriculus; CB, cibarium. Scale bar: 100 μm. (C) Detail of the fly virtually sectioned along a transversal plane through the thorax. DVM-I, dorsal–ventral muscles; SGs, salivary glands. Scale bar: 100 μm. (D) Detail showing the direct flight muscles DFM49–DFM56. DFM52 is only rudimentarily visible, because it is clipped by the viewing plane. Scale bar 40 μm.
The raw optical sections underlying Figure 3
, and additional virtual sections from two directions orthogonal to the recording plane were used for manual segmentation (Figures 4
A–C). Different organs were marked in different colours. From the segmented planes we generated a 3D model and superimposed it on a digitally reconstructed radiography of the fly (Figures 5
A,B). Figures 5
C,D show the DLMs in dark blue. Laterally, three further groups of DVMs (DVM-I, DVM.-II, DVM-II) are shown in light blue. The DFMs are coloured in yellow. The brain, the CN, and the ThAGl, being the fundamental parts of the CNS, are marked in three dissimilar green tones. Above the ThAGl parts of the intestinal system are shown in red and brown colours. The cibarium (CB) crosses the brain, leading to the oesophagus (ES), which connects to the proventriculus (PV). It is followed by the gut, and the unpaired asymmetrically located crop (CR).
Figure 4. Cross sections in three orthogonal directions (A–C) were used to segment anatomical structures in three different orientations. ES, oesophagus; CN, cervical connective; DLMs, dorsal longitudinal muscles; DVM-I/-II/-III, dorsal–ventral muscles; PV, proventriculus; TDT, tergal depressor of the trochanter; ThAGl, thoracico-abdominal ganglion. I: Sagittal plane, II: transversal plane, III: coronal plane. (A) Sagittal optical slice. (B) Computed coronal slice. (C) Computed transversal slice. Length of scale bars 100 μm.
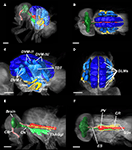
Figure 5. Segmentation of Drosophila organs. (A,B) Overview of the entire fly, illustrating major components of the nervous system, the intestinal tract, and the musculature. (C,D) Detail of the flight musculature. DLM, dorsal longitudinal muscles (dark-blue); DVM-I/II/III, dorsal–ventral muscles (blue); DFM49–DFM54, direct flight muscles (yellow); TDT, tergal depressor of the trochanter (light-blue). (E-F) Detail of the CNS and the intestinal system. Brain (dark green), CN, cervical connective (green); ThAGl, thoracico-abdominal ganglion, (light-green); CB, cibarium (light-rose); ES, oesophagus (rose); PV, proventriculus (red); SGs, pair of salivary glands (brown); CR, crop (orange). Length of scale bar 200 μm.
We have demonstrated that ultramicroscopy allows the 3D-reconstruction of the inner anatomy of entire, cleared Drosophila. The time needed for a complete scan of a single fly, including 3D-reconstruction using Amira, is below 30 min. As mechanical slicing of the specimen is avoided, artefacts, such as tissue disruptions or dislocations due to the microtome knife do not occur. 3D-reconstructions of entire cleared Drosophila have previously been performed by McGurk et al. (2007)
using optical projection tomography (OPT), mainly focussing on the IFMs. Our method not only provides a much more detailed 3D-reconstruction of the IFMs and the TDT, but also highlights the organization of the DFMs. The filigree muscles DFM49 up to DFM54 can be clearly visualized. Ultramicroscopy allows resolutions of <10 μm, and can resolve structures down to the size of single dendritic spines (Dodt et al., 2007
). By combining ultramicroscopy with GFP expression (Dodt et al., 2007
), immunolabeling (Jährling et al., 2008
), or lectin-staining (Jährling et al., 2009
) morphological structures not visible by observations in autofluorescent light can be visualized. The instrumentation effort for an ultramicroscopy setup is moderate. Standard software developed for confocal microscopy can be used for 3D-reconstruction.
Ultramicroscopy data can be segmented for further analysis with respect to various anatomical structures. We presented segmentations of the major parts of the nervous system, the musculature and the intestinal tract. While such segmentations on a manual basis presently still are relatively costly in terms of labour, future developments in the field of computational bioimage processing may allow a semi-automatic processing of anatomical structures of interest. A promising approach in this field may be model based segmentation algorithms, which contain some integrated ‘knowledge’ about the general geometry of various anatomical structures and their variability (Peng, 2009
; Heimann and Meinzer, 2009
).
Drosophila is an important model organism for studying the function of genes linked to neuro-degenerative diseases and how these mutations lead to dysfunction (Lu and Vogel, 2009
). Recent development of RNAi libraries allows now the systematic, genome-wide analysis of tissue morphogenesis (Dietzl et al., 2007
). Since these questions are now in the focus of current molecular genetic research, ultramicroscopy may become important as an appropriate tool for rapid scanning of experimentally generated fly mutants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank T. K. Stackhouse for carefully reading this manuscript. This study was supported by SFB391, the Hertie Foundation and the Max-Planck Society.
The Supplementary Material for this article can be found online at http://www.frontiersin.org/systemsneuroscience/paper/10.3389/neuro.06/001.2010/