Optogenetic dissection of neuronal circuits in zebrafish using viral gene transfer and the Tet system
1
Friedrich Miescher Institute for Biomedical Research, Novartis Research Foundation, Basel, Switzerland
2
Biozentrum, Department of Cell Biology, University of Basel, Basel, Switzerland
3
Department of Bioengineering, Stanford University, Stanford, CA, USA
4
Max Planck Institute for Medical Research, Heidelberg, Germany
The conditional expression of transgenes at high levels in sparse and specific populations of neurons is important for high-resolution optogenetic analyses of neuronal circuits. We explored two complementary methods, viral gene delivery and the iTet-Off system, to express transgenes in the brain of zebrafish. High-level gene expression in neurons was achieved by Sindbis and Rabies viruses. The Tet system produced strong and specific gene expression that could be modulated conveniently by doxycycline. Moreover, transgenic lines showed expression in distinct, sparse and stable populations of neurons that appeared to be subsets of the neurons targeted by the promoter driving the Tet-activator. The Tet system therefore provides the opportunity to generate libraries of diverse expression patterns similar to gene trap approaches or the thy-1 promoter in mice, but with the additional possibility to pre-select cell types of interest. In transgenic lines expressing channelrhodopsin-2, action potential firing could be precisely controlled by two-photon stimulation at low laser power, presumably because the expression levels of the Tet-controlled genes were high even in adults. In channelrhodopsin-2-expressing larvae, optical stimulation with a single blue LED evoked distinct swimming behaviors including backward swimming. These approaches provide new opportunities for the optogenetic dissection of neuronal circuit structure and function.
Genetically encoded tools to visualize and manipulate defined neurons (Zhang et al., 2007
; Luo et al., 2008
) open exciting new perspectives for the mechanistic analysis of neuronal circuits (Adamantidis et al., 2007
; Zhang and Oertner, 2007
; Douglass et al., 2008
; Huber et al., 2008
; Arrenberg et al., 2009
; Gradinaru et al., 2009
; Petreanu et al., 2009
; Wyart et al., 2009
). Exploiting the full potential of these tools would be greatly facilitated by small animal models that permit optical access to many individual neurons in vivo. One model that fulfills these requirements is the zebrafish (Baier and Scott, 2009
), which is becoming increasingly more important in neuroscience and other disciplines. In addition, advanced genetic methods are required to express transgenes at high levels in specific cell types. We therefore capitalized on experience in different species to establish gene expression systems in zebrafish that achieve high-level, conditional expression of transgenes in small and specific neuronal subsets.
We first explored whether transgenes can be introduced into zebrafish neurons by viral gene delivery. Although this approach is well-established in mammals (Luo et al., 2008
) it has, to our knowledge, not been explored systematically in the zebrafish brain, despite some encouraging reports (Wagle et al., 2004
).
Alternatively, transgenes can be expressed by generating stable transgenic lines. This strategy is more time-consuming and often produces lower expression levels but allows for non-invasive and highly reproducible transgene expression in defined neuronal subsets. Ideally, a transgenic approach should accomplish three goals. One goal is to generate libraries of transgenic lines with diverse, pseudo-random expression patterns in order to target transgenes to a wide range of cell types. Second, the system should allow for the simple exchange of transgenes without the loss of the specific expression pattern or expression level. A third goal is to regulate the expression level of transgenes by simple pharmacological treatments. Exchangeable transgene expression in a variety of different patterns has recently been achieved in zebrafish by combining transposon-mediated gene- or enhancer-trapping (Kawakami et al., 2004
) and the two-component Gal4-UAS system (Scheer and Campos-Ortega, 1999
; Scott et al., 2007
; Asakawa and Kawakami, 2008
; Distel et al., 2009
). This strategy has been pioneered in Drosophila (Fischer et al., 1988
; Brand and Perrimon, 1993
) and requires two transgenic elements that can be present in independent activator and responder lines. By combining different activator and responder lines, different transgenes can be expressed in a variety of predictable patterns that are determined by the activator lines. Putative limitations of the Gal4-VP16 system such as toxic effects of Gal4-VP16 and lack of pharmacological control over the Gal4-VP16 regulated transgene (Gill et al., 1998
; Scott et al., 2007
; but see Distel et al., 2009
) could potentially be overcome by conditional two-component gene expression systems such as the Tet system (Gossen and Bujard, 1992
; Huang et al., 2005
), the insect-specific ecdysone receptor system (Esengil et al., 2007
), or the LexPR system (Emelyanov and Parinov, 2008
). Because the Tet system is most advanced and used very successfully in mammals (Schonig and Bujard, 2003
; Bockamp et al., 2008
), we explored whether the Tet system can be used for the generation of zebrafish libraries with strong, conditional and sparse transgene expression patterns in the brain.
Our results show that, in zebrafish, the combination of Tet activator and Tet-responder transgenes provides a stable and strong doxycycline (Dox)-regulated expression of the Tet responder transgene. Expression patterns were sparse and biased towards those cell types in which the promoter of the Tet-activator is active. This property of the Tet system can be exploited to express transgenes in small, defined neuronal subsets. Moreover, we show that the high expression level enables efficient optical control of action potential firing by channelrhodopsin-2 (ChR2) using one- and two-photon stimulation. In freely behaving larvae, optical stimulation of ChR2 produced behavioral responses including backward swimming. Transgenic approaches using the Tet system in zebrafish therefore provide a wide range of opportunities for the structural and functional dissection of neuronal circuits.
Transgenic Fish Lines
Zebrafish (Danio rerio) were bred in the laboratory at 24–27°C. Transgenic fish lines were generated by microinjection of DNA into one-cell embryos using the Tol-2 transposon as described (Kawakami et al., 2004
).
For expression under the control of the HuC promoter, a 9.6-kb DNA fragment upstream of the HuC coding sequence (Park et al., 2000
) was used. The itTA coding region was excised by XhoI/XbaI from plasmid pBS.htTAnls (Krestel et al., 2001
) and inserted into the pBS.HuC.pA plasmid (Park et al., 2000
; Higashijima et al., 2003
) linearized with XhoI/XbaI to obtain pBS.HuC:itTAnls. Next, a I-ScI (blunted)/NotI fragment of HuC:itTA was cloned into the Bsp120I/HpaI-linearized pT2AL200R150G (Kawakami et al., 2004
) to obtain pT2A:HuC:itTA.
For expression under the control of the Dlx4/6 promoter, a 6.0 kb Dlx4/6 promoter fragment was used (Zerucha et al., 2000
). The SmaI/SmaI fragment of the itTA coding sequence was inserted into Ecl136II/Ecl136II-linearized plasmid p1229.Dlx4/6:eGFP (Zerucha et al., 2000
) to obtain pBS.Dlx4/6:itTA. Subsequently, a StuI/ApaI fragment of Dlx4/6:itTA was inserted into ApaI/HpaI-linearized pT2AL200R150G (Kawakami et al., 2004
) to obtain pT2A:Dlx4/6:itTA.
To create the Tet-responder plasmid, a NotI/BglII fragment containing the unidirectional TetO7-Ptet minimal promoter driven Venus was excised from pAAV:iCre:Ptet:Venus (Tang et al., 2009
) and cloned into Bsp120I/BglII-linearized pT2AL200R150G (Kawakami et al., 2004
) to obtain pT2A:Ptet:Venus. Next, a BamHI(Blunted)/Bsp1407I-digested ChR2YFP fragment was excised from pLECYT plasmid (Boyden et al., 2005
) and used to replace the EcoRV/Bsp1407I Venus fragment in pT2A:Ptet:Venus to obtain the pT2A:Ptet:ChR2YFP. All plasmids were purified by Phenol/Chloroform. mRNA for pCS2FA-transposase was prepared by in vitro transcription of the Tol2 transposase ORF contained in pCSTP plasmid (Kawakami et al., 2004
).
To generate stable transgenic fish lines, plasmids pT2A:HuC:itTA (10 ng/μl) or pT2A:Dlx4/6:itTA (10 ng/μl), pT2A:Ptet:ChR2YFP (10 ng/μl) and CSTP mRNA (20 ng/μl) were co-injected into one cell embryos. Potential founders (F0) were identified by ChR2 gene specific PCR reactions on genomic DNA. Genomic DNA was extracted from fin-clips using Proteinase-K digestion followed by DNA precipitation. Primers: sChR2: 5′-GAATGAGAATGACCGGGCAG 3′ and antiChR2: 5′-CTAGATACAGCATGGACGGGTTC-3′ for ChR2; itTA FP: 5′-GTTGGCATTGAGGGCTTGAC-3′ and itTA RP: 5′ TCCTTGGCAACTTGGTGCTC-3′ for itTA. PCR-positive founders were crossed to obtain double positive, fluorescent larvae, which were selected and raised up to adulthood as F1-founders. Different founder lines were established after two generations of out-crossing with wild type to avoid multiple integrations.
Recombinant Viruses
To construct rAAV virus, either a 2.8-kb HuC promoter-driven eGFP minigene (NotI/SalI) previously shown to mimic the 9.6 kb HuC promoter (Park et al., 2000
) or a bi-directional Ptet minimal promoter driving iCre and mCherry (pAAV:iCre:Ptet:mCherry) (Tang et al., 2009
) was constructed and packaged.
To generate recombinant rSindbis virus, the coding sequence with a pA signal of ChR2YFP (XbaI/ApaI) or AlstR:IRES:eGFP (NheI/PmeI) (Tan et al., 2006
) was cloned in the rSindbis genome linearized by XbaI/ApaI or NheI/PmeI, respectively. The internal ribosomal entry sequence (IRES) sequence used here contains only 10 AUGs and an A7 (AAAAAAA) bifurcation loop, while optimal IRES sequences contain 12 AUGs and an A6 (AAAAAA) bifurcation loop (Bochkov and Palmenberg, 2006
; Kwan et al., 2007
). As a consequence, the second gene in the bicistronic construct will be expressed with low efficiency. Recombinant viruses were produced as previously described (Ehrengruber, 2002
; Tan et al., 2006
; Wickersham et al., 2007
). Viruses were titrated by estimating fluorescent cells in dissociated neuronal cultures. Titers obtained were ∼1012 particles per ml for rAAV, ∼109 particles per ml for rSindbis:ChR2YFP, ∼107 particles per ml for rSindbis:AlstR:IRES:eGFP, ∼108 particles per ml for rRabies:eGFP, and ∼108 particles per ml for rRabies:mCherry. All animal procedures were performed in accordance with official animal care guidelines and approved by the Veterinary Department of the Canton of Basel-Stadt (Switzerland).
Imaging and Electrophysiology
Confocal imaging was performed using an Olympus Fluoview microscope with two excitation lasers (488 nm and 543 nm) and emission filters for eGFP/YFP (505/50 nm) and RFP (570-nm long pass). Two photon microscopy was performed as previously described (Yaksi and Friedrich, 2006
) using a customized two-photon microscope equipped with a 20× water immersion objective (NA 0.95 or 1.0; Olympus or Zeiss) and a Ti:Sapphire laser (SpectraPhysics, Mountain View, CA, USA). Fluorescence emission was detected externally by a photomultiplier-based whole-field detector through an emission filter (GFP and YFP: 535/50; mCherry: 640/75). For optical stimulation of ChR2 with blue light, a light-emitting diode (LED; Philips Lumileds, San Jose, CA, USA; 470 nm) was attached to the lamp port of the epifluorescence condenser. For 2-photon activation, the laser was scanned across the sample using a conventional frame scan pattern. Laser intensity was regulated to zero during flyback. For behavioral experiments, a single LED (Philips Lumileds, San Jose, CA, USA) equipped with a collimator was positioned approximately 5 cm away from the Petri dish at an angle of approximately 45°.
Electrophysiological experiments were performed in an explant preparation of the intact zebrafish brain (Friedrich and Laurent, 2001
; Yaksi and Friedrich, 2006
) in teleost ACSF (Mathieson and Maler, 1988
). Whole-cell patch clamp recordings were performed using borosilicate micropipettes (8–18 MOhms) and a Multiclamp 700B amplifier (Molecular Devices). Intracellular pipette solution contained (in mM) 130 Kgluconate, 10 Na-gluconate, 10 Na phosphocreatine, 4 NaCl, 4 Mg-ATP, 0.3 Na-GTP, and 10 HEPES (pH 7.25). Neurons expressing ChR2YFP were identified and targeted using a combination of multiphoton and transmitted light microscopy.
Doxycycline Administration
To test conditional regulation of gene expression by Dox, larvae (10-20 per group) were raised in 90-mm Petri dishes at 28.5°C in ∼50 ml of filtration-sterilized standard embryo medium containing (in mM): 5 NaCl, 0.17 KCl, 0.33 CaCl2, and 0.33 MgSO4. Dox was added to a final concentration of 20 ng/ml and the medium was refreshed every 8 h. Adult zebrafish (2–4 fish per group) were kept at 27°C in 1-l tanks without water circulation containing 50 μg/ml Dox in 0.8 l of filtration-sterilized fish water. Control fish were kept under the same conditions in the absence of Dox. After 5 days of Dox treatment, one group of fish was inspected under a fluorescence microscope (larvae) or sacrificed for in situ hybridization (adult). Another group was returned to the fresh medium or circulating system containing no Dox and analyzed 4–6 days (larvae) or 6 days (adults) later.
In Situ Hybridization
The ChR2YFP-specific probe was cloned by PCR into a pCRII-TOPO TA cloning vector (Invitrogen) using two oligos: 5′-ATGGATTATGGAGGCGCCCT-3′ (sChR2), and 5′ AAGTGGCGCCCCACACAATT-3′ (antiChR2). Antisense and sense probes were synthesized by in vitro transcription using DIG RNA Labeling Kit (SP6/T7) (Roche Applied Science). In situ hybridization was performed as described (Geisen et al., 2008
). Briefly, brains were dissected and fixed in 4% PFA overnight, dehydrated, and stored at −20°C in 100% methanol. Hybridization was carried out at 70°C overnight in 50% formamide, 5 × SSC, 1% SDS, 100 μg/ml tRNA, 50 μg/ml heparin. After extensive wash, the brains were blocked with Blocking Reagent (Roche Applied Science) in MABT (pH 7.5; 100 mM maleic acid, 150 mM NaCl, 0.1% Tween 20). Brains were incubated overnight at 4°C with an anti-DIG antibody conjugated to alkaline phosphatase (1/5,000; Roche Diagnostics). After several washes, the alkaline phosphatase activity was detected using NBT (nitro-blue tetrazolium chloride; 250 μg/ml) and BCIP (5-bromo 4 chloro-3-indolyl phosphate; 180 μg/ml) diluted in a solution of (in mM) 100 NaCl, 100 Tris (pH 9.5), 50 MgCl2, 2 levamisole and 0.1% Tween. Brains were then fixed in 4% PFA and stored at 4°C for further analysis.
Stereotactic Microinjection of Recombinant Viruses
Adult fish were anesthetized by immersion in 0.01% tricaine methanesulfonate (MS-222) and placed into a custom-made holder. Anesthesia was maintained by continuous superfusion of the gills with MS-222-containg fish water via a cannula inserted into the mouth. A small craniotomy (diameter, ∼80 μm) was made above the target region in the forebrain using a dentist drill. A glass micropipette (diameter, ∼15 μm) containing recombinant virus in PBS was inserted through the craniotomy into the target area using a motorized manipulator and approximately 50–100 nl of the solution was injected by pressure. The coordinates of target areas (OB, Vv, Dp or lateral telencephalon) were calculated according to the zebrafish brain atlas (Wullimann et al., 1996
). To examine expression of transgenes, fish were sacrificed 2, 3, 4, 14 or 90 days after virus injection.
For virus injection in larvae, fish were anesthetized by immersion in MS-222 and embedded in 2% low-melting agarose. A micropipette containing recombinant virus in PBS was inserted through the skin and cartilage into the target area under visual control. Expression was assayed at various time points after virus injection by MS-222 anesthesia, embedding in low-gelling agarose, and 2-photon microscopy.
Behavioral Analysis
Individual zebrafish larvae were placed in a 35 mm Petri dish and filmed at a rate of 30 ms/frame using a video tracking system (Zebralab, Viewpoint, France). A light-emitting diode (LED; Luxeon V-Star; 470 nm) equipped with a collimator was placed next to the Petri dish at a distance of approximately 5 cm and an angle of approximately 45°. The LED was driven using a commercially available driver (Luxdrive BuckPuck, LEDdynamics) and produced ∼0.35 mW/mm2 at the location of the dish. Each larva was tested only once and acclimated to the Petri dish for 2–3 min before the trial was started. A trial consisted of three 20-s periods. Blue light was off during the first and the third period and on during the second period. Swimming trajectories were extracted using Viewpoint software and further analyzed in Matlab. Swimming speed was quantified as the mean displacement between adjacent frames, divided by the frame time, and binned into 1-s bins. The on-response was quantified as the mean change in swimming speed during 2 s after light onset relative to a 10-s baseline period before light onset. The off-response was quantified as the mean change in swimming speed during 20 s after light offset relative to a 10-s baseline period before light offset.
Viral Gene Delivery
We tested ten recombinant viruses for their potential to infect neurons in the brain of larval and adult zebrafish: recombinant adeno-associated virus (rAAV) serotypes 1, 2, 5, 6 and their binary mixtures (1/2, 1/5, 1/6), lentivirus, Sindbis virus and Rabies virus (Furuta et al., 2001
; Luo et al., 2008
). In pilot experiments, viruses containing genes for fluorescent proteins were injected into the larval or adult forebrain. In our hands, rAAV or lentiviruses did not produce detectable fluorescence in the brain, although fluorescence in the skin around the injection site was observed after injection of rAAV (data not shown). Sindbis and Rabies viruses, in contrast, produced strong fluorescence signals in neurons (Figure 1
).
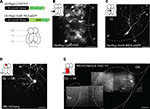
Figure 1. Viral gene delivery. (A) Top: rSindbis constructs with ChR2YFP or AlstR:IRES:eGFP reporters. Bottom: schematic outline of the adult zebrafish forebrain (ventral view). (B,C) Fluorescence images of neurons expressing ChR2YFP (B) or AlstR:IRES:eGFP (C) 12–24 h after injection of Sindbis virus (2 photon microscopy; z-projections of stacks). Insets indicate injection sites and visualized areas. Titer of injected virus was high in (B) and low in (C). (D) All or none expression of mCherry in OB mitral cells after injection of Rabies virus in a target area (Dp), indicating retrograde tracing. (E) Visualization of an axonal trajectory from Vv to Dp in a HuC:YC transgenic fish. Background fluorescence is from YC. Scale bars: 50 μm.
We further characterized gene delivery by Sindbis viruses using eGFP or a fusion of the light-gated cation channel Channelrhodopsin-2 (ChR2) and YFP (ChR2YFP) (Nagel et al., 2003
; Boyden et al., 2005
) as reporters (Figure 1
A). Fluorescence was observed in neurons as early as 12 h after virus injection and became very strong after 24 h in larvae and adults (Figure 1
B). To better assess the expression level, we replaced the reporter gene by a bicistronic construct consisting of the allatostatin receptor (AlstR) gene, an IRES, and the eGFP gene (AlstR:IRES:eGFP) (Tan et al., 2006
). In zebrafish, the reporter gene after the IRES sequence is expressed with relatively low efficiency (Kwan et al., 2007
). Here we used a suboptimal IRES that decreases expression levels even further (Bochkov and Palmenberg, 2006
). Indeed, injection of plasmid DNA containing AlstR:IRES:GFP under the control of a strong neuronal promoter (HuC) (Park et al., 2000
) did not produce detectable fluorescence in embryos (data not shown). When delivered by Sindbis virus, however, the same recombinant gene, AlstR:IRES:eGFP, produced robust fluorescence (Figure 1
C), demonstrating that expression levels achieved by Sindbis infection are higher than those obtained by conventional transgenesis.
We next analyzed expression produced by a modified Rabies virus that does not cross synapses because the Rabies glycoprotein was replaced by the red-fluorescent protein mCherry (Wickersham et al., 2007
). Fluorescent neurons were detected 3–4 days after virus injection and persisted for at least 3 weeks. mCherry-positive neurons appeared very bright (Figures 1
D,E), consistent with the all-or-none expression expected from viral replication within infected cells (Wickersham et al., 2007
). Targeted injection of Rabies virus into the dorsal posterior telencephalon (Dp), a remote telencephalic target area of the olfactory bulb (OB), labeled mitral cells in the OB (Figure 1
D) and neurons in the ventral nucleus of the ventral telencephalon (Vv) (Figure 1
E), consistent with results obtained by non-viral tracing techniques (Rink and Wullimann, 2004
). When projections remained superficial, individual axons could be followed completely by multiphoton microscopy in the intact brain of adult fish (Figure 1
E). Hence, modified Rabies virus infects zebrafish neurons and functions as a very bright neuronal tracer, consistent with observations in mammals (Wickersham et al., 2007
).
We next analyzed combinatorial transgene expression in zebrafish using the E. coli-derived Tet system consisting of (i) the transactivator tTA (Tet-repressor-VP16) and (ii) the tTA- responder element Ptet (Gossen and Bujard, 1992
; Zhu et al., 2002
; Huang et al., 2005
; Luo et al., 2008
). In the Tet-Off configuration, tTA binds to Ptet and initiates transcription of the Ptet-regulated transgene. The Tetracycline derivative Dox binds to tTA and dramatically decreases its affinity to Ptet. As a result, Ptet remains unoccupied and transcription at Ptet is turned off (Gossen and Bujard, 1992
; Luo et al., 2008
). In the upgraded version of tTA used in this study (improved tTA; itTA) (Baron et al., 1997
; Krestel et al., 2001
; Tang et al., 2009
) the Tet-repressor gene was codon optimized and the VP16 domain of tTA replaced by FFF domains to avoid cytotoxicity (Gill et al., 1998
; Scott et al., 2007
). In the mouse brain, a unidirectional Ptet responder construct showed different expression patterns, possibly due to positional effects of the Ptet responder transgene insertion (Mayford et al., 1996
). We therefore used a unidirectional Ptet element to obtain different expression patterns in different lines.
To test for toxicity, variability and regulation of our Tet system components we first generated lines harboring two transgenes: (i) a construct containing itTA under the control of a 9.6 kb zebrafish HuC promoter fragment (Park et al., 2000
), and (ii) a ChR2YFP responder controlled by Ptet (Figure 2
A). In constitutive transgenics harboring a single promoter-reporter construct, expression driven by the HuC promoter is pan-neuronal at early larval stages (Park et al., 2000
) but becomes restricted in adults. In the adult OB, expression is limited to axons of afferent sensory neurons and mitral cells (Li et al., 2005
) (Figure 2
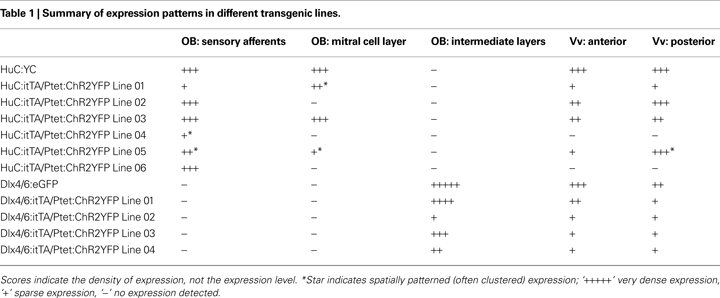
B). Compound HuC:itTA/Ptet:ChR2YFP transgenic fish showed robust YFP-fluorescence in the larval and adult brain. Contrary to conventional transgenics, however, ChR2YFP expression patterns varied substantially between independent founder lines (Table 1
). Out of six founder lines analyzed as adults, one (Line 03) showed transgenic expression similar to conventional HuC promoter-transgenic fish (Figures 2
C,D), while the remaining lines showed strong ChR2YFP expression in different, sparse populations of neurons (Figures 2
E–I). The ChR2YFP expression patterns of the founder lines differed already at larval stages and, within each line, remained stable through several generations (Figures 2
C–H). In Line 02, for example, ChR2YFP was expressed in afferent sensory axons projecting to multiple glomeruli but not in mitral cells. Very similar expression patterns were observed in multiple individuals from different generations (Figures 2
E,F). In the OB of Line 05, ChR2YFP was expressed in a subpopulation of afferent sensory axons projecting to the lateral OB and to a prominent glomerulus in the ventral OB, while medial projections were not labeled (Figures 2
G,H). In addition, ChR2YFP was expressed in a small set of mitral cells located in the medial OB, while no mitral cells were observed in the lateral OB. The number of mitral cells observed in confocal image stacks (300 μm in z-dimension) was very similar in different individuals (mean ± SD: 9.3 ± 1.5 MCs; n = 10 OBs in five fish). In none of the founder lines did we observe expression in deep layers of the adult OB where inhibitory interneurons are located or in other brain areas that do not exhibit expression in conventional HuC promoter transgenics (Table 1
). Hence, different HuC:itTA/Ptet:ChR2YFP transgenic lines expressed ChR2YFP in distinct and well-defined subpopulations of neurons that appear to be subsets of the neurons targeted by the HuC promoter in constitutive HuC transgenic fish.
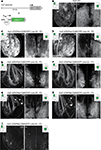
Figure 2. Expression patterns in Tet system transgenics. (A) Constructs used to express the transactivator (itTA) under control of the HuC promoter and ChR2YFP under Ptet control. (B) Expression of the cytosolic fluorescent protein, YC, under the control of the HuC promoter in a transgenic fish generated by conventional techniques. Expression is shown in two forebrain regions of an adult fish (OB and Vv; ventral view; z-projections of small confocal stacks; regions are indicated in insets). Variation among different founder lines is minimal (not shown). In the OB, expression is restricted to afferent sensory axons (open arrowheads) terminating in glomeruli (open arrows) and mitral cells (filled arrowheads). In Vv, expression is seen in distributed neurons along the midline. (C–I) Expression patterns in Tet system transgenics. Genotype, line number and generation (F0: injected generation) are indicated. Unless noted otherwise, images were taken with the same settings to compare brightness. Expression patterns vary between founder lines but are stable between generations. (C,D) Offspring of HuC:itTA/Ptet:ChR2YFP Line 03 strongly express in sensory afferents (open arrowheads), glomeruli (open arrows), mitral cells (filled arrowheads) and Vv neurons. Expression in the OB was similar to HuC:YC transgenics (B). Laser power in (D) was 10 % of that used for other images. (E,F) Offspring of HuC:itTA/Ptet:ChR2YFP Line 02 express selectively in subsets of sensory afferents and in Vv neurons. (G,H) Offspring of HuC:itTA/Ptet:ChR2YFP Line 05 express in subsets of sensory afferents projecting to the lateral OB and to a ventral glomerulus (open arrow), in a small group of mitral cells in the medial OB (filled arrowheads), and in a dense population of neurons in posterior Vv. (I) Offspring of HuC:itTA/Ptet:ChR2YFP Line 04 express in a sparse set of sensory afferents projecting to the posterior-lateral OB. Scale bars: 50 μm.
Sparse and distinct Ptet:ChR2YFP expression patterns were also obtained when we used the Dlx4/6 promoter for itTA expression (Zerucha et al., 2000
) (Figure 3
A). In the OB, the Dlx4/6 promoter directs gene expression to a subset of GABAergic interneurons mainly in intermediate layers, presumably periglomerular cells. Afferent sensory axons or mitral cells do not express transgenes under the control of the Dlx4/6 promoter (Li et al., 2005
) (Figure 3
B). In different Dlx4/6:itTA/Ptet:ChR2YFP lines we observed ChR2YFP fluorescence in distinct subsets of neurons that were reproducible for at least three generations (Figures 3
C,D and Table 1
). Out of four Dlx4/6:itTA/Ptet:ChR2YFP lines analyzed in detail as adults, one line expressed ChR2YFP in many neurons in the intermediate layers of the OB (Figure 3
C), similar to the expression pattern of constitutive Dlx4/6:eGFP transgenics (Figure 3
B). In the three other lines, expression was limited to sparse subsets of neurons. In Line 02, for example, fluorescence was restricted to a small number of neurons with relatively large somata and extensive dendritic arborizations (Figure 3
D). In all Dlx4/6:itTA/Ptet:ChR2YFP lines, no labeling of mitral cells or afferent sensory axons was observed and expression was restricted mainly to the intermediate layers (Table 1
). Thus, results obtained with two different promoters and a Ptet:ChR2YFP responder construct indicate that transgenesis using the Tet system with the unidirectional Ptet promoter can generate a variety of stable and sparse expression patterns that can be restricted to selected cell types by the promoter used for itTA expression.
Figure 3. Expression patterns in Tet system transgenics using a second promoter. (A) Constructs used to express the transactivator (itTA) under the control of the Dlx4/6 promoter and ChR2YFP under Ptet control. (B) GFP expression in the adult OB of Dlx4/6:eGFP transgenic fish generated by conventional methods. Expression is observed in interneurons in the intermediate layers. (C) Offspring of Dlx4/6:itTA/Ptet:ChR2YFP Line 01 express in many OB interneurons. (D) Offspring of Dlx4/6:itTA/Ptet:ChR2YFP Line 02 express in a sparse subset of OB interneurons. Scale bars: 50 μm.
We noticed that the fluorescence in Tet system transgenics was very strong. Fluorescence images from the OB of HuC:itTA/Ptet:ChR2YFP transgenics (Figure 2
C) were brighter than images from a conventional HuC:YC line when acquired using the same settings (Figure 2
B), even though YC is a very bright cytosolic protein whereas ChR2YFP is a transmembrane protein. To further evaluate expression levels, we replaced ChR2YFP by the bicistronic AlstR:IRES:eGFP and found that in HuC:itTA/Ptet:AlstR:IRES:eGFP, the GFP fluorescence was still strong despite the weak IRES element used for the translation initiation of eGFP (data not shown). The Tet system therefore affords enhanced expression compared to conventional approaches using endogenous promoters, presumably because the itTA-induced expression acts as an amplifier of the promoter used for itTA expression.
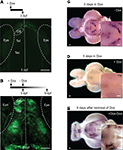
When HuC:itTA/Ptet:ChR2YFP (Line 03) embryos were raised in the presence of 20 ng/ml Dox in the water, no fluorescence was detectable at 3-days post fertilization (dpf) (Figure 4
A), whereas siblings raised in the absence of Dox exhibited strong fluorescence (data not shown). After removal of Dox, fluorescence appeared 4 days later and reached control levels within approximately 6 days (Figure 4
B). In adults (Figure 4
C), expression analyzed by in situ hybridization became undetectable 5 days after addition of Dox (50 μg/ml) to the water (Figure 4
D) and returned to control levels 6 days after removal of Dox (Figure 4
E). Fluorescence levels decreased slowly and became undetectable only after >2 months, indicating that the ChR2YFP fusion protein is very stable in mature neurons. Importantly, zebrafish treated with 100 μg/ml Dox for up to 1 month showed no signs of toxicity and produced normal offspring, demonstrating that the Tet system enables efficient regulation of gene expression in zebrafish without fatal side effects.
Figure 4. Regulation of gene expression by Doxycycline. (A) Raising larvae in Dox (20 ng/ml) suppresses reporter expression in HuC:itTA/Ptet:ChR2YFP transgenics (Line 03) at 3 dpf. Weak fluorescence anterior to the OB is autofluorescence in the skin. (B) Strong expression resumed 6 days after Dox removal. (C) Detection of ChR2YFP mRNA by whole mount in situ hybridization in the brain of an adult zebrafish (HuC:itTA/Ptet:ChR2YFP Line 03; ventral view; OB removed). A strong signal was detected in the hypothalamus (Hp; enlarged in inset). (D) In situ signal after 5 days of Dox treatment (50 μg/ml). (E) In situ signal 6 days after removal of Dox. OB: olfactory bulb; Tel: telencephalon; Tec: optic tectum. Scale bars: 100 μm.
In order to explore the Tet system for optogenetic approaches we first performed targeted whole-cell recordings from neurons expressing ChR2YFP under Ptet control in an explant preparation of the adult zebrafish brain (HuC:itTA/Ptet:ChR2YFP Line 03; n = 6). Sustained wide-field optical stimulation (1 s) using a blue LED (470 nm; total power at sample ∼0.3 mW; average power per area ∼0.25 mW/mm2) evoked a pronounced depolarization and action potential firing that adapted over time (Figure 5
A). In voltage clamp, a similar adaptation was observed in the light-evoked inward current (Figure 5
B). These results are consistent with the known properties of ChR2 in neuronal cell culture (Boyden et al., 2005
).
Figure 5. Optical manipulation of neuronal activity in Tet system transgenics expressing channelrhodopsin-2. (A) Response of a mitral cell to prolonged illumination with blue light in current clamp. (B) Response of a mitral cell to blue light stimulation in voltage clamp. (C) Responses of a mitral cell to trains of light pulses (15 ms) at constant frequencies. Other mitral cells showed very similar responses (n = 3). (D) Number of action potentials (mean ± SD) evoked by regular trains of light pulses as a function of frequency, averaged over n = 3 mitral cells. (E) Jitter of action potential timing (mean ± SD) as a function of frequency, averaged over n = 3 mitral cells. (F) Responses to Poisson trains of light pulses (15 ms). Top: example of an intracellular recording. Middle: action potentials (black ticks) evoked by the same stimulus train (blue ticks) in five repeated trials in the presence of kynurenic acid (KYN; 2 mM), which suppresses spontaneous activity (see membrane potential trace on the right). Bottom: Mean firing rate as a function of time. Spike trains were convolved with a Gaussian kernel (sigma: 100 ms) and averaged. Gray shading indicates SD; dashed blue trace shows event rate of the pulse train. Other mitral cells showed very similar responses (n = 3). (G) Responses of the same mitral cell as in (F) in the absence of kynurenic acid, which results in a background of high spontaneous activity (right).
Neurons in the OB exhibit a high rate of spontaneous sub and suprathreshold activity that depends on glutamatergic input (Tabor and Friedrich, 2008
). In order to measure the precision of action potential firing evoked by ChR2 stimulation, we suppressed spontaneous activity by blockade of glutamate receptors with kynurenic acid (2 mM) or by cutting the olfactory nerve and measured responses to trains of short (5–15 ms) blue light pulses. Individual light pulses at a constant frequency triggered single action potentials that faithfully followed the stimulus without failures for frequencies up to 20 Hz (Figures 5
C,D; n = 3 mitral cells). The jitter of spike times was on the order of a few milliseconds and increased only slightly with stimulus frequency (Figure 5
E). Reproducible temporal firing patterns were also observed in response to trains of blue light pulses with Poisson-distributed inter-pulse intervals (Figure 5
F; n = 3 mitral cells). We then examined whether temporal structure can also be imposed on mitral cell firing patterns in the intact network when spontaneous activity is not suppressed. The temporal structure of the light stimulus was clearly observed in mitral cell spike trains, despite the high rate of spontaneous activity (Figure 5
G). Transgenesis using the Tet system does therefore not appear to compromise the function of neurons and enables ChR2-mediated optical control of neuronal activity in the intact brain.
Fine optogenetic manipulations of neuronal circuits require the ability to manipulate action potential firing in individual ChR2-expressing neurons embedded in a larger population. This is difficult to achieve by activation of ChR2 with blue light because the light beam may also depolarize fibers of passage and neurons outside the focal plane. A potential solution to this problem is multiphoton activation of ChR2, which has been used successfully to evoke action potentials in cultured neurons (Rickgauer and Tank, 2009
). In intact tissue, however, action potentials evoked by multiphoton stimulation of ChR2 have, to our knowledge, not been reported. In fact, scanning a mode-locked Ti:Sapphire (multiphoton) laser beam with >150 mW power over hippocampal neurons expressing ChR2 under the synapsin-1 promoter caused depolarizations of only ≤ 1.5 mV and failed to evoke action potentials (Zhang and Oertner, 2007
). Moreover, high excitation power dramatically degrades the spatial resolution of ChR2 activation due to the long lifetime and consequent saturation of the conducting state (Rickgauer and Tank, 2009
). One way to overcome this problem is to increase the density of ChR2 molecules by high expression levels. We therefore tested whether action potentials can be evoked by 2-photon activation of ChR2 at low laser power in HuC:itTA/Ptet:ChR2YFP or Dlx4/6:itTA/Ptet:ChR2YFP transgenic fish.
We recorded in whole-cell mode from ChR2-expressing OB interneurons (Dlx4/6:itTA/Ptet:ChR2YFP Line 01; n = 5) while a region around the neuron was scanned with a mode-locked Ti:Sapphire laser at modest intensities (<10 mW; often <1 mW; 880 nm). The input resistance of the recorded neurons was between 0.4 and 2.4 GOhms (mean ± SD: 1.2 ± 0.9 GOhms; n = 5) and the spread of the focus in z was ∼5 μm (FWHM of Gaussian fit). In all neurons, scanning caused large-amplitude (>10 mV) rhythmic depolarizations at the scanning frequency that triggered one or multiple action potentials (Figure 6
A). In voltage clamp, 2-photon stimulation evoked pronounced inward currents (Figure 6
B). In current clamp, action potentials were evoked with 100% probability by the first stimulus frame. Failures to evoke action potentials occurred only occasionally during later frames (Figure 6
A). Similar observations were made in MCs (n = 2) and Vv neurons (n = 4).
Figure 6. Multiphoton activation of channelrhodopsin-2 in Tet system transgenics. (A) Frame scanning at low laser power evokes a strong depolarization and action potential firing (current clamp). (B) Frame scanning evokes inward currents (voltage clamp). (C) Currents evoked by frame scanning as a function of focus position.
In order to assess the z-resolution of ChR2 activation, we measured currents evoked by 2-photon stimulation at different focal planes above and below the recorded soma. Currents were largest when the focus was at the level of the soma and decreased rapidly when the focal plane was moved 10–20 μm away (Figure 6
C). The current decayed to 1/e of the maximum current when the focal plane was displaced from the soma by approximately 5 μm in the z direction. Hence, activation of ChR2 was confined to a narrow plane, as predicted for ChR2 activation at low laser power (Rickgauer and Tank, 2009
). Expression of ChR2 under Tet control therefore enables efficient and spatially precise control of action potentials by multiphoton excitation.
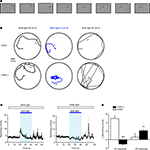
We next examined whether blue light stimulation with an external LED can evoke behavioral responses in freely swimming larvae expressing ChR2YFP under Tet control. HuC:itTA/Ptet:ChR2YFP fish (Line 03) were outcrossed to wild-type fish to produce ChR2 positive and ChR2-negative siblings. Individual larvae were then placed in a 35 mm Petri dish under normal room light and exposed to a constant blue light stimulus (LED; 470 nm; ∼0.35 mW/mm2) for 20 s. ChR2-negative larvae responded to the onset of the blue light stimulus with a brief increase in swimming speed (n = 30). Subsequently, swimming patterns appeared normal under blue light. During a period of approximately 20 s after stimulus offset, larvae occasionally darted through the dish but otherwise swam normally. Eighty percent (24/30) of ChR2-positive siblings, in contrast, responded to the onset of the blue light stimulus with a transient period of slow backward swimming (∼2 s; Figure 7
A; Movie S1 in Supplementary Material; n = 30). After this unusual behavior, ChR2-positive larvae swam forward again but their average speed was slightly reduced and their trajectories appeared to be more confined than those of ChR2-negative siblings (Figures 7
B,C). Following the offset of the blue light, ChR2-positive larvae also darted frequently (Figure 7
B). The same assay was also performed with other wt lines (Ab/TÜ or TL; n = 38 larvae) and with Dlx4/6:itTA/Ptet:ChR2YFP (Line 01) larvae (n = 12). None of these fish showed backward swimming in response to blue light.
Figure 7. Optical modulation of swimming behavior in larvae expressing channelrhodopsin-2 under Tet control. (A) Video frames taken immediately after onset of blue light (LED; inter-frame interval: 300 ms). Note that the larva (HuC:itTA/Ptet:ChR2YFP Line 03) swims backwards (see also Movie S1 in Supplementary Material). (B) Examples of swimming trajectories of individual larvae (ChR+: HuC:itTA/Ptet:ChR2YFP Line 03; ChR-: wild-type sibling) during 20-s periods before, during and after exposure to blue light. Arrows indicate direction of swimming. (C) Mean swimming speed of ChR- and ChR+ larvae before, during and after a 20-s exposure to blue light (n = 30 for each group). (D) Quantification of on-response (mean change in swimming speed during 2 s after light onset) and off-response (mean change in swimming speed during 20 s after light offset) in ChR- and ChR+ larvae (n = 30 for each group). **P < 0.01; ***P < 0.001 (paired t-test).
To quantitatively compare the behavior of ChR2-positive HuC:itTA/Ptet:ChR2YFP (Line 03) larvae and ChR2-negative siblings, we first measured the average swimming speed in 1-s time bins (Figure 7
C). We then calculated the change in swimming speed during the first 2 s after light onset (‘on-response’) and during 20 s after light offset (‘off-response’) relative to the preceding 10 s baseline periods. In ChR2-positive larvae, the on-response was significantly smaller than in wt siblings (paired t-test; P < 0.001; n = 30 larvae in each group; Figure 7
D). Indeed, the average swimming speed decreased, rather than increased, relative to the baseline period because backward swimming is slower than the average spontaneous swimming speed. The off-response of ChR2 positive larvae was significantly larger than that of wt siblings (paired t test; P < 0.01; n = 30 larvae in each group; Figure 7
D). These results demonstrate that ChR2 expression under Tet control permits optical manipulation of behaviors in zebrafish larvae using a remote LED.
Our results demonstrate that spatial and temporal control of gene expression in zebrafish can be achieved by viral gene delivery and the Tet system. Whereas viruses are fast to generate and easy to apply, the Tet system allows for the conditional, non-invasive expression of transgenes in defined cell types. The high expression levels obtained with both approaches are critical for many applications such as the use of IRES sequences or the optogenetic manipulation of neuronal activity and behavior. Hence, viral gene delivery and the Tet system are interesting genetic tools for efficient and specific manipulations of neurons in zebrafish.
Transgene expression in neurons was observed upon injection of two recombinant viruses, Sindbis and Rabies, with complementary advantages. Sindbis virus allows for an easy exchange of inserts and fast, high-level gene expression in neurons but the time window for functional experiments is limited to 1 or 2 days in mammals (Furuta et al., 2001
). Rabies virus, on the other hand, is more difficult to modify but causes little or no cytotoxicity for at least 2 weeks in mammals (Wickersham et al., 2007
). Moreover, Rabies virus is taken up by axons and functions as a bright retrograde neuronal tracer (Wickersham et al., 2007
). In addition, we observed infection of skin cells by rAAV, indicating that gene delivery by rAAV is, in principle, possible in zebrafish. A previous study reported that zebrafish neurons can be infected by baculovirus (Wagle et al., 2004
). Viral vectors are therefore likely to provide a rich repertoire of gene delivery tools in zebrafish neurons that has not yet been exploited.
The Tet system provides several advantages over other conditional gene expression systems in zebrafish (Esengil et al., 2007
; Emelyanov and Parinov, 2008
) including (1) minimal side effects on gene regulation (Reboredo et al., 2008
), (2) no detectable toxicity of the modulator (Dox), (3) no interference with hormone signaling, and (4) the availability of optimized components that have been tested extensively in other species (Gossen and Bujard, 1992
; Baron et al., 1997
; Tang et al., 2009
). Moreover, the recent development of caged Dox provides the opportunity to fine tune the control of gene expression using optical approaches (Cambridge et al., 2009
).
In addition, we found that different transgenic lines generated with the same Tet system constructs expressed the transgene in a variety of unique and stable patterns. This phenomenon is reminiscent of the diverse expression patterns in thy-1 promoter-transgenic mice (Caroni, 1997
; Feng et al., 2000
) and can be used to generate libraries of sparse, cell type-specific expression patterns. One potential reason for the diversity of expression patterns could be influences of the genomic environment onto the short, unidirectional responder element (Mayford et al., 1996
) but further studies are necessary to explore the underlying mechanisms. Importantly, expression patterns in Tet system transgenics were not random but restricted to cell types included in the expression spectrum of the promoter driving itTA. Compared to other approaches for the generation of expression libraries such as the thy-1 promoter or the Gal4/UAS system, the Tet system thus provides an additional layer of genetic control that can be exploited to pre-select cell types of interest. The Tet system therefore combines several useful features of a conditional gene expression system in zebrafish.
Multiphoton activation of ChR2 expressed under Tet control with low laser power produced large inward currents that easily triggered action potentials in different types of neurons. This high efficiency was most likely due to the high expression levels achieved by the Tet system. In general, low levels of ChR2 expression cannot simply be compensated by higher excitation power because this degrades the effective spatial resolution of optical stimulation, both for single- and for multiphoton activation of ChR2 (Schoenenberger et al., 2008
; Rickgauer and Tank, 2009
). High expression levels as obtained by the Tet system are therefore important for precise optogenetic manipulations in complex circuits.
Low intensities of blue light evoked behavioral responses in freely swimming larvae expressing ChR2 under Tet control. The combination of high-level gene expression and a small, transparent model organism thus permits the manipulation of neuronal activity in behaving animals without the need for surgical implantation of optical fibers. Interestingly, blue light stimulation evoked backward swimming in HuC:itTA/Ptet:ChR2YFP (Line 03) larvae, a behavior that we have never observed during spontaneous swimming. Backward swimming is slower than free forward swimming or escape swimming and does not involve large-amplitude tail beats, indicating that it is not simply the reverse of a motor program for forward swimming. Since blue light stimulation did not evoke backward swimming in wt fish and in a transgenic line expressing ChR2YFP in a different pattern (Dlx4/6:itTA/Ptet:ChR2YFP Line 01), backward swimming is most likely triggered by the activation of a specific set of neurons. Different swimming patterns are known to be controlled by command centers in the hindbrain (McLean and Fetcho, 2008
; Orger et al., 2008
) or by neurons in the spinal cord (Wyart et al., 2009
). A first step towards identifying the neuronal circuitry controlling backward swimming would thus be the spatial mapping of neuronal stimulation to motor output in HuC:itTA/Ptet:ChR2YFP (Line 03) larvae, which may be achieved by local optical stimulation of neurons in different brain regions (Arrenberg et al., 2009
). The diversity of conditional expression patterns and the high expression levels produced by the Tet system in zebrafish therefore open interesting perspectives for studies of neuronal circuit structure and function.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank E. Callaway for AlstR:IRES:eGFP, K. Kawakami for Tol2 transposon materials, R. Malinow for help with Sindbis virus and all Friedrich lab members for discussions. This work was supported by the Novartis Research Foundation, the Deutsche Forschungsgemeinschaft (SFB 780), the Boehringer Ingelheim Fonds, the Swiss National Fonds, the Max Planck Society and the VW foundation.
The Supplementary Material for this article can be found online at http://www.frontiersin.org/neuralcircuits/paper/10.3389/neuro.04/021.2009/
Park, H. C., Kim, C. H., Bae, Y. K., Yeo, S. Y., Kim, S. H., Hong, S. K., Shin, J., Yoo, K. W., Hibi, M., Hirano, T., Miki, N., Chitnis, A. B., and Huh, T. L. (2000). Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 227, 279–293.
Zerucha, T., Stuhmer, T., Hatch, G., Park, B. K., Long, Q., Yu, G., Gambarotta, A., Schultz, J. R., Rubenstein, J. L., and Ekker, M. (2000). A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 20, 709–721.