Abstract
Increasing public awareness of foodborne illnesses, factory farming, and the ecological footprint of the meat industry, has generated the need for animal-free meat alternatives. In the last decade, scientists have begun to leverage the knowledge and tools accumulated in the fields of stem cells and tissue engineering toward the development of cell-based meat (i.e., clean meat). In tissue engineering, the physical and biochemical features of the native tissue can be mimicked; cells and biomaterials are integrated under suitable culture conditions to form mature tissues. More specifically, in skeletal muscle tissue engineering, a plurality of cell types can be co-cultured on a 3D scaffold to generate muscle fibers, blood vessels and a dense extracellular matrix (ECM). This review focuses on tissue engineering of skeletal muscle and the adjustments needed for clean meat development. We discuss the skeletal muscle structure and composition, and elaborate on cell types from farm animals that have the potential to recapitulate the muscle ECM, blood vessels, muscle fibers and fat deposits. We also review relevant biomaterials, primarily for fabricating scaffolds that can mimic the intramuscular connective tissues, as well as gene expression studies on the biological pathways that influence meat quality.
Introduction
Why Clean Meat?
Clean meat, meat produced from cells cultures, is a prominent alternative for traditional meat, derived from live animals (Post, 2012; Slade, 2018). This approach gained increasing attention in public opinion, popular media, animal welfare organizations, the scientific community, and among investors (Goodwin and Shoulders, 2013; Schneider, 2013; Verbeke et al., 2015; Stephens et al., 2018b), particularly after the production of the first clean meat prototype (Post, 2014). Animal-based food products are considered inefficient, as animals consume large amounts of food throughout their lives, of which up to 97% of the calories are lost for processes regarding body maintenance and the production of non-edible tissues (Schmidinger, 2012; Flachowsky et al., 2018; Gaydhane et al., 2018; Sarlio, 2018). When compared to other industries, animal-based products have a larger environmental footprint compared to plant-based products in terms of soil and water demand, and greenhouse gas (GHG) emission, with the beef industry imparting the heaviest environmental impact (Peters et al., 2007; Tuomisto and de Mattos, 2011; Eshel et al., 2014; Scarborough et al., 2014). According to the report of the Food and Agriculture Organization of the United Nations, the livestock sector is responsible for 14.5% of GHG emissions, and taps 30% of Earth's terrain and 8% of the global freshwater (Steinfeld et al., 2006; Gerber et al., 2013). With an expected doubling of the global demand for meat by 2050, traditional meat production systems cannot be considered sustainable.
Intensive factory farming and poor animal welfare conditions are common causes of foodborne illnesses such as swine and avian influenza (Greger, 2007). They also contribute to the spreading of E.coli, salmonella and campylobacter, which can be found in meat (Anomaly, 2015). Producing meat in a sterile and controlled environment could prevent these problems and improve food safety. In addition, 70–80% of the antibiotics used in the United States are given to farm animals (Kümmerer, 2003; Elliott, 2015). Overuse of antibiotics induces selection of antimicrobial resistant (AMR) strains, a major human health concern (Avesar et al., 2017; Chen et al., 2017; Tang et al., 2017). Colistin AMR originated in 2015 in pig farms (Reardon, 2017) and was later found in chicken and pig in Vietnam and South America (Nguyen et al., 2016; Monte et al., 2017). It is estimated that by 2050, AMR will be responsible for more deaths than cancer (O'Neill, 2014) and to cost $2.9 trillion annually to the OECD countries (Friedman et al., 2016). Although the magnitude of antibiotics use in clean meat production still requires investigation, it has the potential to be lower than the current use in the meat industry due to the sterile culture conditions and the possible use of antibiotic-free media (Specht et al., 2018).
Ethical concerns regarding factory farms (Verbeke and Viaene, 2000; Schröder and McEachern, 2004; Herzog, 2016) and the slaughter of animals is another drive behind clean meat research (Hopkins and Dacey, 2008; van der Weele and Driessen, 2013; Sharma et al., 2015). The public and the scientific community have come to respect farm animals as sentient beings, with physical and psychological needs (Hughes, 1995; Dawkins, 2006; Duncan, 2006; Webster, 2006; Panskeep and Others, 2012). The cost of raising a chicken humanely (Broom, 1991; Broom and Fraser, 2015), similar to that of a pet (Council, 2010), is 2–3 orders of magnitude higher than the cost of products made from that animal (United States Department of Agriculture, 2018). In other words, animal-based meat must compromise on animal interests, due to economic constraints (Webster, 2001; Lusk and Norwood, 2011). Since the overwhelming majority of domesticated animals are raised for meat, pushing for the obsoletism of animal-based meat is essential from an altruistic stance (Green, 2008; Elder and Fischer, 2017; Broad, 2018).
From a culinary perspective, fine-tuning of meat components is difficult in live animals, and is achieved by selecting specific breeds and following complex feeding protocols (Smith et al., 2009). In clean meat, cells are cultured in-vitro in a customizable and controlled platform. The simple and precise manipulation of the cell microenvironment could be used to fine-tune meat traits. In addition, combinations of cells from different species may give rise to new and unprecedented flavors. This simple, fast and intuitive meat manipulation platform can facilitate a new R&D field of customizable meat products.
A New Objective
An engineered tissue is traditionally optimized for biological functionality, improved post-transplantation viability, biodegradability and negligible immune response (Lanza et al., 2011). To reach these goals, quantifiable protocols are developed, and a rigorous series of experiments are performed to assess the effect of specific factors on the overall quality of the tissue. Optimally, experiments are conducted using factorial designs, in which both main factors and their interactions are measured (Chen et al., 2011). A similar approach is required for clean meat optimization, with focus on flavor (taste and aroma), texture, cost, nutritional value and food safety (Miller et al., 2001; Edelman et al., 2005; Post and van der Weele, 2014; Listrat et al., 2016; Gasteratos and Sherman, 2018).
Flavor assessments are a key challenge in clean meat research. Meat contains thousands of flavor molecules (Pearson and Dutson, 1994; Shahidi, 2012), including amino acids, hemoproteins (Nollet, 2012), sulfur and carbonyl compounds (Calkins and Hodgen, 2007), lipids (Shahidi, 2002; Wood et al., 2004), short peptides (Claeys et al., 2004), and additional flavor volatiles (Shahidi et al., 1986; Gorraiz et al., 2002). An additional hurdle stems from the small size of lab samples; standard protocols used to analyze meat quality are designed to test samples ranging from whole animals (Mitchell, 2007) to tens of grams (Anderson, 2007), which are at least three orders of magnitude larger than experimental lab samples. Light diffraction is a fast, inexpensive and non-invasive meat analysis method (Prieto et al., 2009; Damez and Clerjon, 2013). This technology is based on a reference database of light diffraction patterns from meat samples with known composition. The correlation between a measured sample and the reference database is used to estimate the sample's composition with high accuracy. Scaling down this technology is feasible (Jordan et al., 2009) and could accelerate clean meat research. However, light diffraction only provides an estimate of main components, such as fat and collagen content in the sample. Molecular profiling of lab samples can be obtained by chromatographic separation, followed by mass spectrometry identification and quantification (Shahidi, 2012; Trivedi et al., 2016). A reference database of molecular profiles obtained from meat samples could be beneficial to interpret and standardize experimental results (D'Alessandro et al., 2012; Trivedi et al., 2016; Gilbert-López et al., 2017). While molecular profiling may be slower and more expensive, it provides quantification of flavor molecules, and is more suited for developing novel tissues, which composition may often be outside the scope of the reference database.
Texture measurements are simpler, and can easily predict sensory attributes (Luckett et al., 2014). They can be performed using existing devices (Kramer and Szczesniak, 1973; Pearson and Dutson, 1994), but may require adaptation of current protocols or instruments to assess smaller samples in broader suboptimal ranges. Factors that can influence clean meat texture include muscle fiber maturation (Koohmaraie et al., 2002) and alignment, extracellular matrix (ECM) protein content and alignment, intramuscular fat (IMF) content (Kramer and Szczesniak, 1973), and scaffold material and structure. Static strain can be used to increase myotube and ECM alignment with the added value of improved muscle maturation (De Deyne, 2000; Edelman et al., 2005; Post, 2012; Heher et al., 2015); however, the scalability of the process may be difficult. Scalable alignment can be induced via scaffold design, by generating aligned fibrous scaffolds or by micropatterning (Choi et al., 2008; Aviss et al., 2010; Rodriguez and Larkin, 2018).
Cost is probably the key hurdle, as clean meat borrows tools from the expensive and heavily regulated field of regenerative medicine, and applies them in the food industry, which has one of the lowest margins of profit. The main expenditures of clean meat production are biomaterials and their purity assurance, human resources, facilities and measurement tools. Specht (2019) estimates that 55–95% of the production cost could be attributed to media use, and that 99% of the media cost can be attributed to Growth factors. Her review discusses different scenarios for cost reduction. While growth factors (GFs) are costly biomaterials, due to protein purification (Van der Gucht, 2018), their contribution to tissue engineering is invaluable. Replacing purified GFs with GF-producing cells is possible, however may complicate the regulation of GF concentration, as experimental parameters can influence GF expression levels in these cells. Two alternative approaches are to produce these molecule at a lower grade of purification (or without purification), or to generate inexpensive analogs of these molecules. Customized bioreactors need to be developed and optimized for clean meat production, to improve the efficiency of media use, and recycle main media components (Moritz et al., 2015; Specht et al., 2018; Stephens et al., 2018a).
Cells
Meat, or skeletal muscle, contains about 90% muscle fibers, 10% connective and fat tissues (Listrat et al., 2016) and 0.3% blood (Warriss and Rhodes, 1977; Nollet, 2012). Skeletal muscle tissues, aimed for tissue regeneration, can be generated in 3D scaffolds by co-culturing muscle cells, endothelial cells (ECs) and ECM-producing supporting cells (Figure 1) (Edelman et al., 2005; Levenberg et al., 2005; Shandalov et al., 2014; Gholobova et al., 2015; Egozi et al., 2016), but lack fat and blood components. In order to recapitulate the fat composition of the native skeletal muscle tissue, IMF should be generated from adipocytes. As for blood components, with the exception of fish, the majority of the hemoproteins in meat stem from myoglobin (on average 1.5% of the protein content of meat) found inside muscle fibers (Listrat et al., 2016). This means that the production of “clean blood” might be of low concern, especially since blood cultures are not established yet for medical purposes. Neural cells also play a crucial regulatory role in the skeletal muscle, providing electrical signals that influence tissue homeostasis and maturation (Langelaan et al., 2011; Post and Hocquette, 2017). While innervation may be difficult to attain in a scalable and cost-effective manner, electrical signals may mimic some of the neural cells essential roles (Langelaan et al., 2010). While current methods are considered wasteful for clean meat production, it may be advantageous to explore more energy efficient means of artificial electrical stimulations. As for the cell source, the following sections will emphasize the use of bovine cells, as developing clean beef could be considered as a pragmatic approach for the initial stages of clean meat production, since (1) the overwhelming majority of tissue engineering research is done in mammalian cells, (2) beef is considered an expensive meat, (3) beef has a larger ecological impact compared to other conventional meats (Tuomisto and de Mattos, 2011), and (4) biopsies from larger animals are less invasive. Biomarkers of major cell types found in bovine muscle have been previously summarized (Du et al., 2015; Guo et al., 2015). Additional cell types found in the muscle, their roles and biomarkers were summarized by Tedesco et al. (2017).
Figure 1
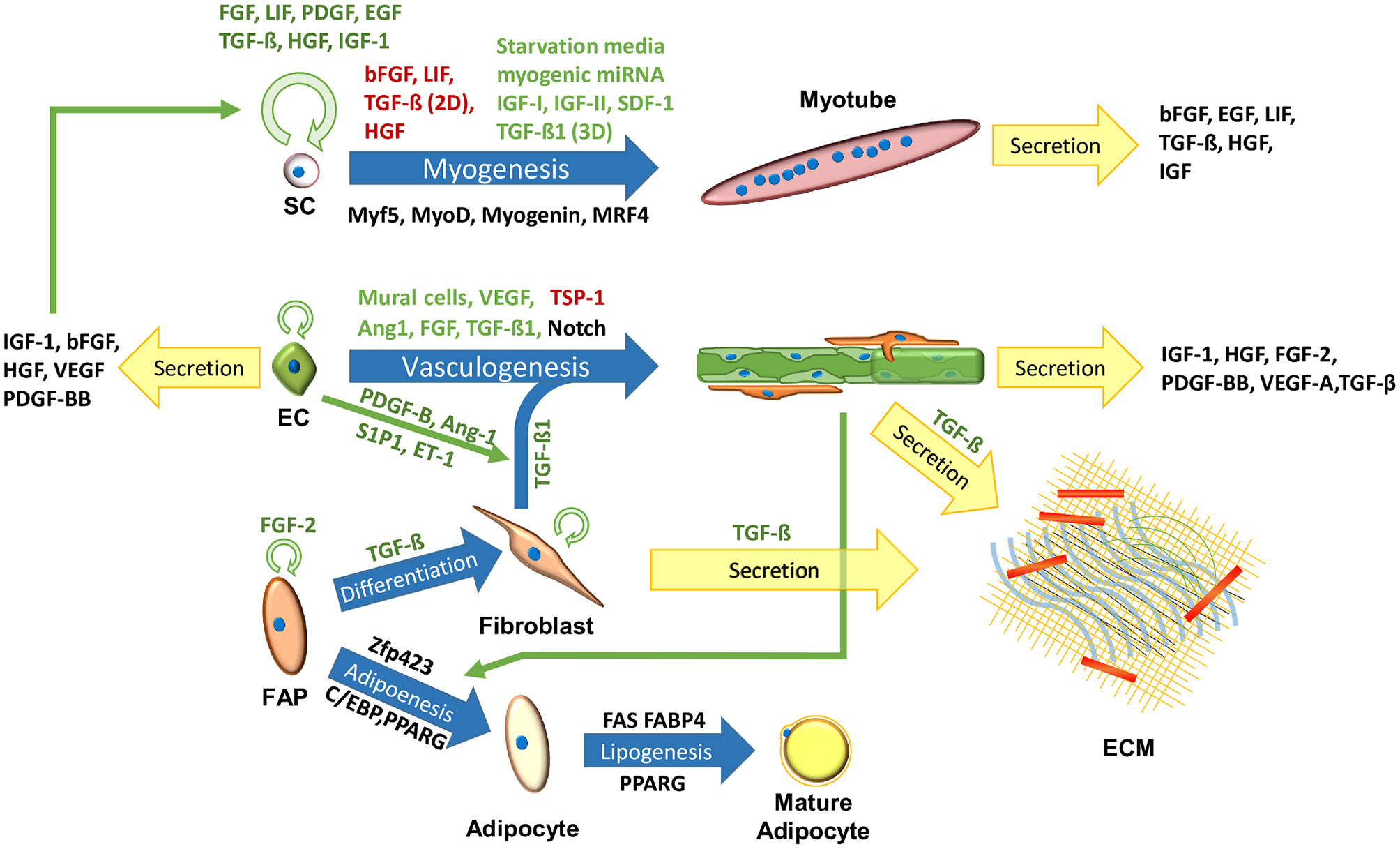
Cells in the skeletal muscle niche. Blue arrows signify maturation processes. Black, green, and red text next to the arrows signify key biomarkers, inhibitors and enhancers of the process, respectively. Circular green arrows signify cell proliferation.
Stem Cells
Stem cells are a population of progenitor cells, which can proliferate to increase the cell population, and differentiate to develop a specialized functionality. Stem cell research is an established niche in animal science, focused on myogenic and adipogenic stem cells, crucial for clean meat research (Dodson et al., 2015). A recent report described a first stable culture of bovine embryonic stem cells (ESC) (Bogliotti et al., 2018; Yuan, 2018), which could potentially differentiate into all cell types required to recapitulate skeletal muscle development (Du et al., 2010a; Thorsteinsdóttir et al., 2011; Yan et al., 2013; Chal and Pourquié, 2017), and form a cell bank with an unaltered and stable karyotype, eliminating further dependence on animals for cell isolation. Mesenchymal stem cells (MSC) are another promising candidate, due to their abundance and roles during muscle development (Du et al., 2010b), and their ability to differentiate into myocytes, adipocytes, fibroblasts and ECs (Oswald et al., 2004; Bosnakovski et al., 2005; Huang et al., 2012; Yan et al., 2013; Jana et al., 2016; Ramírez-Espinosa et al., 2016; Okamura et al., 2018). During muscle development, the majority of MSCs commit to the myogenic lineage, generating muscle fibers and a pool of satellite cells. Other MSCs commit to the fibro/adipogenic lineages, generating a pool of fibro/adipogenic progenitor cells (FAPs) (Du et al., 2013). MSCs undergo senescence in-vitro (Bonab et al., 2006; Wagner et al., 2008), however, under appropriate culture conditions, certain MSC subtypes can expand and maintain their differentiation potential (King and Miller, 2007; Nekanti et al., 2010; Cardoso et al., 2012; Gottipamula et al., 2016). Induced pluripotent stem cells (iPSC) can also be considered as a cell source for clean meat production. iPSC are simple to produce and are extensively studied, making them valuable for lowering the cost and research needed to develop stable stem cell lines. They also simplify the cell isolation, allowing to harvest ubiquitous matured cell types (Keeney et al., 2012). However, they require gene editing, suffer from low yields, and may not precisely mimic the behavior primary stem cells (Roberts et al., 2015; Specht et al., 2018). As such, it is possible they would complicate clean meat regulatory procedure and consumer acceptance.
Satellite Cells and Myogenesis
Satellite cells (SCs) are adult stem cells, which can be found in skeletal muscle tissues, and take part in muscle repair (Péault et al., 2007; Cosgrove et al., 2009; Yin et al., 2013). Methods for working with SCs extracted from different species have been summarized by Burton et al. (2000). The paper provides a table of references sorted by species, and a comparison of SC characteristics, response to GFs, and isolation, growth, and differentiation protocols. Bovine satellite cells (BSCs) were used to create the first clean meat prototype (Post, 2014), and their growth in bioreactors for clean meat production was recently assessed (Verbruggen et al., 2018). BSCs can be isolated from bovine carcasses (Dodson et al., 1987; Frey et al., 1995; Muroya et al., 2001; Kamanga-Sollo et al., 2004, 2008; Ding et al., 2018), biopsies (Frey et al., 1995) or fetuses (Bridge et al., 1998; Will et al., 2015). The muscle tissue is ground and enzymatically digested using pronase, collagenase II or trypsin. The BSC fraction is then enriched by a preplating method and can be further purified by FACS of cells positive for CD56 and CD29, and negative for CD45 and CD31 (Ding et al., 2018).
SC proliferation and myogenesis can be regulated by the judicious use of GFs (Figure 1). Addition of LIF (Vitello et al., 2004; Nicola and Babon, 2015), TGF-β (Vitello et al., 2004; Kollias and McDermott, 2008) and FGF (Allen and Boxhorn, 1989; Pawlikowski et al., 2017) to the BSC media in 2D cultures, has been shown to inhibit spontaneous differentiation, crucial for optimal cell expansion. Addition of LIF (Austin and Burgess, 1991; Nicola and Babon, 2015), heparin-binding epidermal growth factor (hb-EGF) (Thornton et al., 2015), TGF-β (Vitello et al., 2004) and insulin-like growth factor 1 (IGF-1) (Allen and Boxhorn, 1989; Vitello et al., 2004) can improve BSC proliferation. In addition to its central role in muscle development, TGF-β contributes to mature skeletal muscle mass, and is a key regulator of intramuscular fibrogenesis (Kollias and McDermott, 2008; Miao et al., 2016). In addition, it was shown that TGF-β1 suppressed myogenesis in 2D cultures, while enhancing myogenesis in 3D cultures, rendering it a valuable regulator of in-vitro muscle tissue development (Krieger et al., 2018). Myogenesis can be induced by substrate stiffness (Levy-Mishali et al., 2009; Syverud et al., 2014), IGFs and myogenic miRNA (Vitello et al., 2004; Yin et al., 2013), or by inhibition of anti-myogenic GFs produced by nearby cells (Christov et al., 2007; Yin et al., 2013). Inexpensive analogs for key GFs, or of additional molecules that can influence their metabolic pathways [e.g., myogenic miRNA (Ge et al., 2011)], may benefit clean meat research (Ding et al., 2018).
A detailed list of SC biomarkers, including the lineage markers, Pax7 and Myf5, was summarized by Yin et al. (2013). These and additional biomarkers can be used to track SC differentiation into myotubes (Zammit et al., 2006). The predominant test to quantify SC differentiation is the fusion index (FI), which is defined as the percentage of nuclei inside the myotubes from the total number of nuclei in the sample. FI can be determined by assessing (1) myotube-specific nuclear staining (myogenin), (2) colocalization of non-specific nuclear staining (e.g., DAPI) and antibodies that stain proteins abundant in myotubes (e.g., MYH and desmin), or (3) inexpensive colorimetric stainings (Veliça and Bunce, 2011). Methods 1 and 2 can also be used to quantify 3D samples, and method 2 also be used to visualize myotube morphology. It should be noted that the main role of BSCs is in muscle repair, and that they are formed in later stages of muscle development (Du et al., 2015). BSCs also undergo senescence in-vitro, which might be minimized by recapitulating the SC niche (Boonen et al., 2009; Cosgrove et al., 2009; Yin et al., 2013; Roberts et al., 2015; Jana et al., 2016; Cai et al., 2017; Ding et al., 2018).
Fibroblasts and ECM
The ECM, i.e., the acellular part of the tissue that surrounds the cells, is a key component of all tissues. The skeletal muscle ECM is responsible for tissue elasticity, generates adhesion points for cell adherence, provides cells with a 3D environment and regulates cell proliferation, differentiation, migration, morphology and alignment via biochemical and biophysical cues (Tse and Engler, 2011; Hausman, 2012; Chaturvedi et al., 2015; Fuoco et al., 2016; Grzelkowska-Kowalczyk, 2016). The ECM composition and roles are not constant, but change during the muscle development in a process called fibrogenesis, in parallel to myogenesis and adipogenesis (Thorsteinsdóttir et al., 2011; Yan et al., 2013; Miao et al., 2016). In addition to its biological roles, the ECM contains nutrients such as proteins (mostly collagens), and glycosaminoglycans [mostly hyaluronic acid (HA)], and affects the tissue texture and the overall quality of the meat (Purslow, 2005; Guo et al., 2015; Duffy et al., 2016; Listrat et al., 2016; Huang et al., 2017).
The ECM is mainly composed of collagen fibers and proteoglycan (PG) matrix, but also includes glycosaminoglycans (GAGs) and fibrous proteins such as elastin, fibronectin, and laminin (Thorsteinsdóttir et al., 2011; Listrat et al., 2016). The muscle ECM has three ascending layers of connective tissues, in term of size and wall thickness (Figure 2): the endomysium that fills the gaps between the muscle fibers, the perimysium (Passerieux et al., 2006) that compartmentalize bundles of muscle fibers (fascicles), and the epimysium that envelops the entire muscle (Purslow, 2010; Brazile et al., 2017). The perimysium varies in size and can be divided into primary and secondary perimysium. The primary perimysial layers are smaller, contain fewer muscle fibers and have thinner walls compared to the secondary perimysium (54.6–133 μm wall thickness in bovine) (Purslow, 2010). While all layers are mainly composed of collagens I and III, their relative ratios differ, and other collagen types can be found interspersed. Collagen IV is an essential component of the basement membrane and plays a role in cell-matrix interactions and SC niche (Gillies and Lieber, 2011; Cai et al., 2017). Age-related changes include an increase in collagen concentration, and changes in collagen type distribution and crosslinking (Kragstrup et al., 2011). A thorough review of the different species of collagens, their biological role and tissue distribution was published by Kadler et al. (2007). To a certain extent, mimicking the epimysium, perimysium and endomysium morphology and mechanical properties can be achieved by careful scaffold design. The basement membrane may be difficult to recapitulate prior to cell seeding, as it is tailored to the myotube structure.
Figure 2
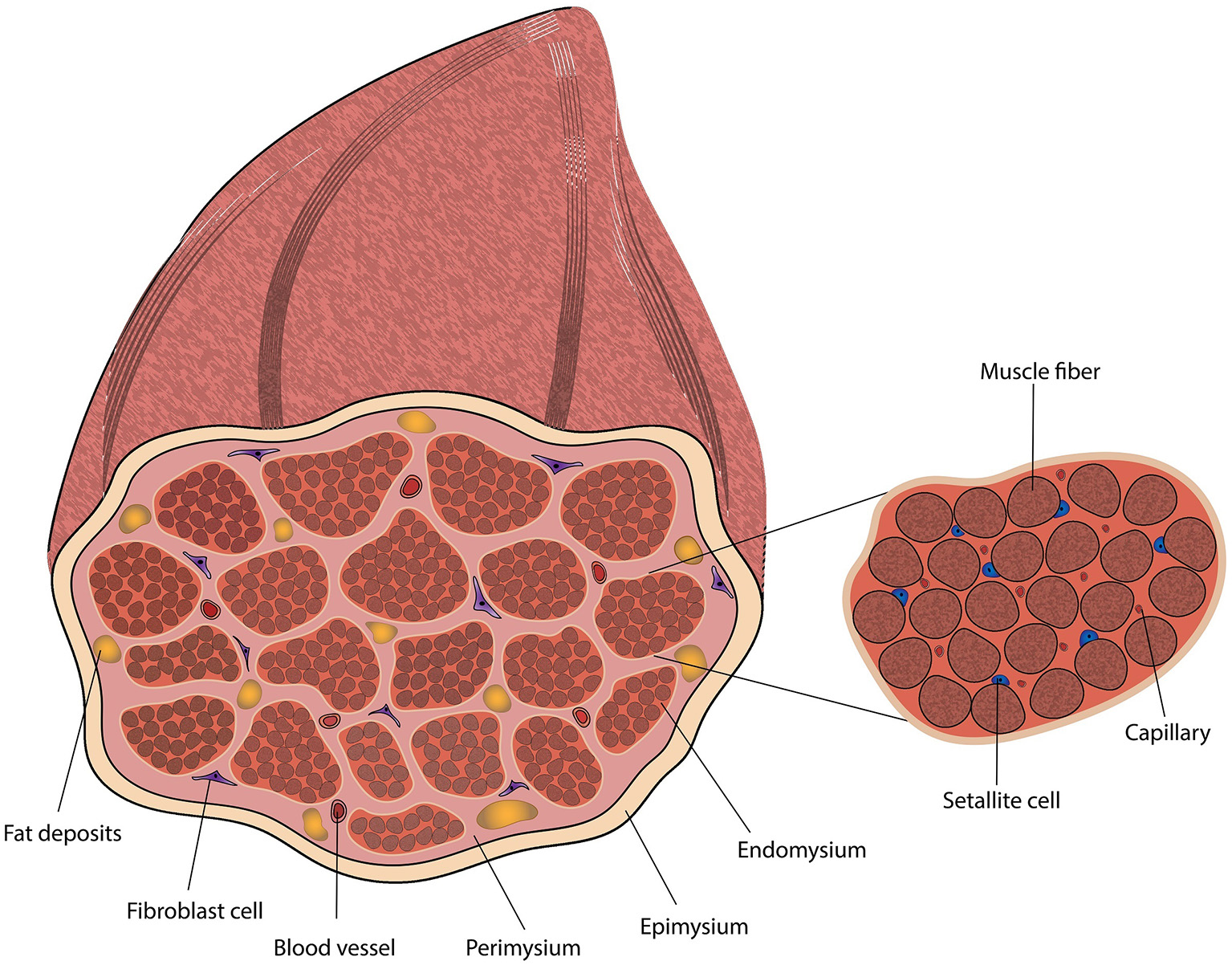
Diagram of skeletal muscle tissue, including a magnification of a muscle fascicle.
Although most cell types contribute to ECM development (Guo et al., 2015), a supporting cell type whose main role is to secrete ECM, is essential. The majority of skeletal muscle tissue ECM is deposited and remodeled by fibroblasts (Figure 1) (Kjaer, 2004; Gillies and Lieber, 2011; Mackey et al., 2017). Fibroblast and myofibroblasts (Baum and Duffy, 2011) are simple to isolate and grow, and have a short cell cycle. They were shown to promote vascularization and muscle development in tissue engineered constructs (Shandalov et al., 2014; Ciccone, 2015; Mackey et al., 2017; Krieger et al., 2018), and are considered to have an important role in clean meat production (Pandurangan and Kim, 2015). However, isolated fibroblasts are usually not very well defined, and are often comprised of several subpopulations with few specific markers (Chapman et al., 2016). In addition, although fibroblast short cell cycle may be beneficial for scale-up processes, it may be unfavorable for co-culture, as their relative concentration is difficult to control. Alternatively, it is possible to produce the ECM separately, lyophilize it into a powder (Gilbert et al., 2005), which can then be used to generate scaffolds or hydrogels for cell seeding. However, such reconstituted ECMs will be homogenous, which may fail to fill the functional, geometrical and compartmental roles of the ECM (Gillies and Lieber, 2011; Huang et al., 2017).
ECs and Vascularization
Muscle tissues are rich in microvascular networks (Figure 2) (Jain et al., 2005; Guo et al., 2015). These networks are not only essential to surpass diffusion limitation (Griffith et al., 2005), which may be improved in-vitro using perfusion bioreactors (Rouwkema and Khademhosseini, 2016), but are also associated with the SC niche (Christov et al., 2007) and regulate skeletal muscle tissue regeneration and maturation through angiocrine signaling (Figure 1) (Christov et al., 2007; Butler et al., 2010; Koffler et al., 2011; Rafii et al., 2016). Microvascular networks also play a key role in generating the conditions required for adipogenesis (Varzaneh et al., 1994; Cao, 2007, 2013; Du et al., 2015), and impact the maturation of nearby ECM-secreting cells (Ciccone, 2015; Landau et al., 2018), which further regulate muscle regeneration (Forbes and Rosenthal, 2014). ECs can form these networks under appropriate culture conditions (Lovett et al., 2009), such as co-culture with mural cells (Sims, 1986; Allt and Lawrenson, 2001; Bergers and Song, 2005; von Tell et al., 2006; Rouwkema et al., 2008), fibroblasts (Levenberg et al., 2005; Kunz-Schughart et al., 2006) or MSCs (Landau et al., 2017; Perry et al., 2017b; Freiman et al., 2018). Guo et al. (2015) provide evidence of the role of bovine muscle ECs in ECM synthesis and remodeling, and discuss their regulatory effect on myoblasts, SCs and fibro/adipogenic precursors (FAPs). Recent studies showed that bovine stromal vascular cells (SVC) can be used to promote both angiogenesis and adipogenesis in-vitro (Ma et al., 2018), which can be explained by the heterogeneity of the SVC (Wittmann et al., 2015), meaning cell sorting is essential for scale-up. While the most characterized bovine ECs are the bovine aortic endothelial cells (Jaffe, 2012), their biological role may not be optimal for muscle regeneration. Bovine skeletal muscle microvascular endothelial cells are commercially available, and were shown to generate vascular networks in 3D constructs in-vitro with lumen-like structures (Yang et al., 2001). While vascularization has an important role in tissue development, cultured meat of lower cell plurality may be simpler to produce without them, as blood vessels may not be an essential component of meat taste or texture.
Adipocytes and IMF
IMF accounts for about 80% of the muscle fat, while the other 20% is found inside myofibers; its amount and fatty acid composition have a crucial role in meat quality, affecting meat juiciness, flavor, tenderness and nutritional value (Wood et al., 2008; Hocquette et al., 2010; Hudson et al., 2015). IMF is created via adipogenesis, in which stem cells differentiate into adipocytes, and lipogenesis, in which triglycerides are accumulated inside the adipocytes (Figure 1). Adipogenesis commences with MSCs or fibrogenic/adipogenic progenitor (FAP) cells commitment to the adipogenic lineage, which is regulated by Zfp423 (Huang et al., 2012). The resulting preadipocytes can further differentiate into adipocytes in a process regulated by transcription factors from the CCAAT/enhancer-binding protein (C/EBP) family, and by the receptor they induce, i.e., peroxisome proliferator-activated receptor gamma (PPARG) (Huang et al., 2012; Du et al., 2015). Adipogenesis can be induced using a cocktail of adipogenic-inducing agents, anti-diabetic drugs (Troglitazone and Rosiglitazone), or commercial adipogenic differentiation kits (Grant et al., 2008; Lengi and Corl, 2010; Cardoso et al., 2012; Huang et al., 2012; Ma et al., 2018). However, the use of such chemicals may render the tissue unsuitable for consumption. Mehta et al. (2019) provides a detail protocol for bovine adipogenesis aimed for clean meat production. This publication includes a detail isolation and culture protocol of the SVC, and an efficient fatty acid-mediated protocols for adipogenesis in 2D plates and 3D alginate-based scaffolds. SC and fibroblast differentiation into adipocytes was also demonstrated in-vitro (Hocquette et al., 2010). In the lipogenesis metabolic pathway, triglycerides are synthesized from long-chain fatty acids (LCFAs) and glycerol-3-phosphate (G3P) (Hocquette et al., 2010). Functional proteins, such as fatty acid synthase (FAS), fatty acid binding protein 4 (FABP4) and PPARG, promote adipocyte maturation and may be used as lipogenesis markers (Moseti et al., 2016). Simple protocols for tracking adipogenesis and lipogenesis include, quantitative colorimetric oil red O staining (Kraus et al., 2016), fluorometric Nile red staining, FABP4 immunolabeling (Eom et al., 2018), qPCR of PPARG and FABP4, flow cytometry, or one of many commercially available adipogenesis assay kits (Aldridge et al., 2013).
Generating IMF in-vitro may require intensive research, as current adipose tissue engineering research focuses on fat outside the muscle (Hong et al., 2005; Gomillion and Burg, 2006; Brett et al., 2017) and uses human and mouse models, which protocols are inefficient for ruminant animals (Laliotis et al., 2010; Ma et al., 2018). Research on adipose tissue vascularization using matured ECs and MSCs, revealed a synergetic effect between angiogenesis and adipogenesis (Kang et al., 2009; Wittmann et al., 2015; Volz et al., 2016). The edible HA, a major component of the ECM (Qazi et al., 2015), has been used to generate scaffolds that induced adipogenesis in-vitro (Halbleib et al., 2003). Hausman (2012) discussed the effect of the ECM on adipogenesis in farm animals, suggesting that laminin, collagen V and VI are key ECM components in intramuscular adipogenesis.
Research on IMF is extensive in farm animals, such as bovine (Harper and Pethick, 2004; Takenouchi et al., 2004; Taniguchi et al., 2008; Du et al., 2010b; Romao et al., 2011; Cardoso et al., 2012; Huang et al., 2012; Hudson et al., 2015; Guan et al., 2017; Ma et al., 2018), porcine (Hausman and Poulos, 2004; Zhang et al., 2014; Chen et al., 2018; Won et al., 2018), sheep (Dodson et al., 1997; Tong et al., 2008; Zhou et al., 2017; Pan et al., 2018) and chicken (Qiu et al., 2017; Wang et al., 2017a; Cui et al., 2018; Fu et al., 2018), and have brought to the development of a wealth of isolation, growth, characterization and differentiation protocols.
Biomaterials and Scaffolding
Biopolymers, growth factors, enzymes, and numerous analytical molecules, are extensively used for cell culture and tissue engineering. For clean meat production, these molecules should be derived from inexpensive, animal-free sources. Transgenic plants can be used for cost-effective (Xu et al., 2016), mass production of biomaterials such as collagen (Shoseyov et al., 2013; Shoseyov and Stein, 2017), elastin (Conley et al., 2009), trypsin (Woodard et al., 2003), growth factors and antibodies (Sack et al., 2015; Snell et al., 2015; Xu et al., 2016). Werkmeister and Ramshaw (2012) surveyed recombinant proteins used as tissue engineering scaffolds, including their source, cost and scalability. Wang et al. (2017b) discuss a range of platforms for recombinant collagen production and their challenges, including collagen I and III. The paper claims that current platforms are costly, and generate collagen without the correct post-translational modifications, limiting their biological functionality. However, the human collagen I produced in plants by the Shoseyov lab, proved sufficiently similar to native collagen by the author, and was flagged as an inexpensive, scalable production system. In addition, the paper argues that in-vitro cell-free synthesis might be an inexpensive, highly controlled platform for protein synthesis.
In order to promote tissue development, the plurality of cell types need to be co-cultured inside a 3D scaffold that mimics their natural environment. The scaffold can be composed of either a hydrogel, a macroporous, sponge-like biomaterial or their combination. While the macroporous biomaterial can provide the macrostructure and mechanical support, the hydrogel can provide cells an elastic 3D microenvironment. To some extent, scaffolds should recapitulate the different layers of the skeletal muscle connective tissue. The macroporous biomaterial should recapitulate the mechanical properties of the perimysium, the hardened connective tissue that surrounds the fascicles, while the hydrogel inside the pores should mimic the endomysium.
Polymers that degrade during the tissue culture period can be applied, however, disposal methods of the degraded molecule or their continued presence must be taken into account. The degradation kinetics of such polymers can be tuned using enzymes or by incorporating weak points inside the polymer structure (Keeney et al., 2012). Traditionally, scaffolds are designed to degrade into safe biomolecules which can be assimilated into the patient's metabolic pathways. For clean meat purposes, scaffolds should be composed of biopolymers designed to degrade into molecules with favorable organoleptic properties. This could be achieved by judicious polymer selection, monomer chemistry, and chemical modification of the scaffold. Shit and Shah (2014) provide a thorough list of edible polymers, including hydrocolloids, polypeptides, lipids, synthetic polymers and composite polymers. The paper discusses their molecular functionality, composition and mechanical properties, but does not elaborate on their possible applications in tissue engineering. Comprehensive reviews of the role of biomaterials in human skeletal muscle tissue engineering have been published, with emphasis on polymers used for scaffold fabrication (Keeney et al., 2012; Qazi et al., 2015). The concepts behind their fabrication and design, with the proper selection of flavor-enhancing biomaterials, can be used to generate novel hydrogels and scaffolds for clean meat purposes.
Scaffolds
Macroporous scaffolds are sponge-like, 3D biomaterials that define the dimensions of the tissue and provide it with mechanical support. These scaffolds constitute a large portion of the final product, which would be commonly defined as a hybrid cell-based meat. As such, the development of novel meat-like macroporous scaffolds, with consideration to their flavor, nutritional value and texture is required. For example, depending on their synthesis procedure, algae-based scaffolds may contain flavor molecules that could be beneficial for clean fish but undesirable for clean beef production (Sarkar and Choudhury, 2017; Grahl et al., 2018).
Macroporous scaffolds vary mainly in their architecture, which is governed by fabrication techniques (Loh and Choong, 2013), and in their molecular composition. They are often generated from edible materials, such as proteins, polysaccharides, native ECM and decellularized plants (Daamen et al., 2003; Loh and Choong, 2013; Courtenay et al., 2018; Hickey et al., 2018). Plant protein-based scaffolds are an appealing candidate for clean meat, due to their nutritional value, low cost and cytocompatibility (Reddy and Yang, 2011; Chien et al., 2013; Huang et al., 2018). Scaffold architecture can be defined by pore size distribution, porosity, and pore interconnectivity. Higher scaffold porosity can improve nutrient flow, ECM deposition, cell viability, cell adhesion and cell proliferation (Zeltinger et al., 2001). Rational combination of the scaffold architecture, composition, coating and filler hydrogel, gives rise to mechanical cues that regulate cell behavior. Scaffolds can be further arranged using anisotropic architecture, anisotropic materials or surface patterning, which can improve both scaffold texture and myotube alignment and maturation (Woerdeman et al., 2005; Beier et al., 2009; Qazi et al., 2015; Wolf et al., 2015; Jana et al., 2016; Cai et al., 2017; Oguz et al., 2018).
Specific cell types require different cell niches, and will thus better thrive on scaffolds bearing properties identical to their native niches (Zeltinger et al., 2001). While fibroblasts and myoblasts may be indifferent, to some extent, to the pore size, the optimal pore size for other cell types may vary with the experiment design, scaffold composition and cell source (Levy-Mishali et al., 2009; Loh and Choong, 2013). Engineered vascularized skeletal muscle tissues, comprised of muscle cells, fibroblasts and ECs were reported to develop inside PLLA-PLGA scaffolds with a pore size of 200–600 μm (Shandalov et al., 2015; Perry et al., 2017b). The optimal pore size for vascularization of monocultures on a porous PEG scaffold, was shown to be 160–270 μm (Loh and Choong, 2013). Reports of pore size for adipogenesis include 6–70 μm for murine ESCs (Loh and Choong, 2013), 100–300 μm for rat MSCs (Neubauer et al., 2005), 135–633 μm for rat preadipocytes (Patrick et al., 1999), 400 μm or 50–340 μm for human preadipocytes (Von Heimburg et al., 2003; Hemmrich et al., 2005) and 200–400 μm for human MSCs (Hong et al., 2005). Further study on the effect of pore size on bovine intramuscular adipogenesis is still necessary.
The mechanical cues sensed by cells growing on a scaffold, are generated by complex interactions involving the macroporous biomaterial, the hydrogel or liquid inside the scaffold pores, and the cells themselves (Levy-Mishali et al., 2009; Purslow, 2010). While mimicking the elastic properties of fat tissues (~2kPa in humans) can promote adipogenesis (Rowlands et al., 2008; Young et al., 2013; Ghasemi-Mobarakeh et al., 2015), mimicking the elastic properties of skeletal muscles (~10kPa) can promote myoblast fusion, and MSC differentiation to the muscle lineage (Discher et al., 2005; Engler et al., 2006; Rowlands et al., 2008; Lesman et al., 2011; Lapin et al., 2013; Loh and Choong, 2013; Ghasemi-Mobarakeh et al., 2015; Qazi et al., 2015; Perry et al., 2017b). Generating an intricate scaffold with several spatially organized cell niches, and a set of large pores for media perfusion, can mimic natural microenvironments found in the skeletal muscle and further modulate cell localization and differentiation (Zeltinger et al., 2001; Reilly and Engler, 2010; Park et al., 2011; Tse and Engler, 2011; Loh and Choong, 2013).
Hydrogels
Hydrogels are ubiquitous in the field of tissue engineering, as they can mimic the 3D environment of the ECM. As such, they are designed with the consideration of their mechanical properties, cytocompatibility, mass transport and degradation kinetics (Drury and Mooney, 2003; Redaelli et al., 2017). Hydrogels can be used as a soft scaffold, as a source material for macroporous scaffolds, or as an ECM-like biomaterial that fills voids of the macroporous scaffolds (Drury and Mooney, 2003; Yegappan et al., 2018). For the latter, hydrogel polymerization must occur after cell seeding, using a cell-compatible solution and protocol (Drury and Mooney, 2003; Redaelli et al., 2017). The gelation kinetics and technique must be scalable, and allow for homogenous gelation throughout the construct (Ruel-Gariépy and Leroux, 2004; Redaelli et al., 2017).
Hydrogels can be used to produce bio-artificial muscles (BAMs)—strips of engineered skeletal muscle tissues. These constructs can be produced by seeding satellite cells inside a gel, anchored from both sides to mimic tension exerted by tendons (Post, 2012). Like macroporous scaffolds, the anchor points define the dimensions of the tissue and provide it with mechanical support. Over time, the gel shrinks and tension builds up in the BAM, which promotes myotube fusion and protein synthesis (Post and van der Weele, 2014). Gholobova et al. (2015) optimized BAM vascularization, showing fibrin gel is sufficient to form vascularized BAMs. Post (2014) generated circular muscle strips inside collagen/matrigel around hubs of agarose gel. Ten thousand of these strips were required to produce an 85-gram burger.
In order to recapitulate muscle ECM, hydrogels for clean meat should be made from pre-synthesized ECM, polysaccharides or fibrous proteins. Hydrogels commonly used for skeletal muscle tissue engineering are comprised of biopolymers such as fibrin, collagen, or polysaccharides such as HA, alginate, agarose and chitosan (Drury and Mooney, 2003; Shit and Shah, 2014; Qazi et al., 2015; Wolf et al., 2015).
Fibrin is a naturally occurring fibrous protein that forms blood clots in injury sites. It is biologically designed to be a temporary scaffold for de-novo tissue regeneration in-vivo, which can degrade over days in cell media (Jockenhoevel et al., 2001; van Hinsbergh et al., 2001). As such, it is commonly used as a hydrogel for muscle tissue engineering (Koning et al., 2009). Fibrin gel is generated under the thrombin-mediated enzymatic cleavage of fibrinogen, which can be expressed in CHO cells (Harrysson and Lövgren, 2012; Hirashima et al., 2016).
Collagen and HA are naturally found in the muscle ECM. As such, they can be used to mimic some of the biochemical and biophysical properties of the native ECM, and are also susceptible to remodeling and degradation by the cells, crucial for cell migration and ECM maturation (Drury and Mooney, 2003). Collagen is often used in tissue engineering, as it is the most abundant protein in the body. The fibrous collagens I and III are abundant in the muscle ECM. They serve structural roles in the tissues, as anchoring points for cell adhesion, and facilitate cell migration and tissue development (Sweeney et al., 2008; Fuoco et al., 2016). However, collagen generates planar structures in the ECM (Purslow, 2014), and due to their high tensile strength, collagen hydrogels are mostly used to generate mechanically stable, cell adhesive, macroporous scaffolds (Drury and Mooney, 2003; Kadler et al., 2007). A collagen-fibrin mixture can be used as a hydrogel for muscle regeneration, which has been shown to promote cell proliferation and myotube differentiation (Beier et al., 2009).
HA is a simple GAG ubiquitous in the body and in the muscle ECM. It participates in wound healing and can regulate cell behavior, such as adipogenesis, angiogenesis and tissue organization (Colwell et al., 2003; Halbleib et al., 2003; Allison and Grande-Allen, 2006; Davidenko et al., 2010; Chang et al., 2013). HA hydrogels are cell-compatible, show favorable viscoelastic properties, have high water retention, and can be synthesized in animal-free platforms (Sze et al., 2016). HA-collagen composites show improved mechanical and biological properties, and can be used for adipogenesis scaffolding (Drury and Mooney, 2003; Allison and Grande-Allen, 2006; Davidenko et al., 2010; Chang et al., 2013).
Alginate is an inexpensive seaweed-based polysaccharide which forms hydrogels in the presence of Ca+2. It is composed of two monomers, one of which interacts with Ca+2, making crosslink degree highly tunable (Drury and Mooney, 2003). Since alginate is cell-inert, it is routinely functionalized with RGD peptides that provide anchoring points for cell attachment. Tuning the monomer ratio and RGD concentration can improve myoblast proliferation and differentiation (Rowley and Mooney, 2002). Alginate hydrogels were recently used as a scaffold for bovine adipogenesis (Mehta et al., 2019) While alginate cannot be remodeled or degraded by cells, degradation kinetics can be controlled by modifying its structure (Drury and Mooney, 2003). Alginate-HA composites can improve the regenerative properties of the alginate gel, while providing improved gelation compared to HA alone (Oerther et al., 1999).
Chitosan is an edible glucoseamine polymer, used in skeletal muscle tissue engineering (Qazi et al., 2015; Wolf et al., 2015). It is commonly derived from animals, however, can also be produced from mushrooms (Mesa Ospina et al., 2015). Chitosan provide similar structure to GAGs, and can provide antibacterial properties (Fedorovich et al., 2007), however like alginate, it requires chemical modification to facilitate cell adhesion and biodegradability (Huang et al., 2017). Jana et al. (2013) used chitosan scaffolds to generate uniaxial scaffolds with tuned stiffness, resulting in long (500 μm) and thick (50 μm) mouse myotubes that express MHC. Chitosan biopolymers could also be used to optimize the gel's mechanical properties (Enrione et al., 2013).
Gene Expression
Gene expression tools are often used to study metabolic pathways in farm animals, particularly of IMF. These works are highly valuable for clean meat research, as they are not aimed for regenerative medicine, but rather focus on meat traits, such as marbling (Lee et al., 2010; Sadkowski et al., 2014; Chen et al., 2015; Hudson et al., 2015; Meadus et al., 2018), myogenesis (Wang et al., 2005, 2015; Lee et al., 2014; Dai et al., 2016), and sensory qualities (Bernard et al., 2007; Hocquette et al., 2007; Bongiorni et al., 2016; Tizioto et al., 2016). Research on correlations between gene expression and meat quality has been recently summarized in several publications. Linde (2018) published a table (table 2.5) of bovine genes and their corresponding meat traits. Guo and Dalrymple (2017) elaborated on genes which correlate with adipogenesis, lipogenesis, fatty acid composition, meat tenderness, and the connection between ECM and IMF. Picard et al. (2015) elaborated on proteomics, transcriptomic and metabolomics works conducted in bovine, chicken, and porcine. The paper provides a list of biomarkers for beef tenderness and gene clusters with their corresponding pork qualities. It should be noted that most of these works are performed in meat from whole animals, and that the data found in such complicated biological systems may include additional pathways which are irrelevant for clean meat research. In addition, most of these studies are comparative, with the aim of finding novel genes. As such, key genes with similar levels of expression are left unreported.
Gene expression can be measured on three levels of complexity using qRNA, microarrays and RNA sequencing (RNA-seq). qRNA quantifies several genes of interest and can be used to monitor the expression of key biomarkers when a deep understanding of the system is not required. Ramírez-Espinosa et al. (2016) provide a list of PCR primers designed to assess bovine myogenesis, adipogenesis and stemness. Microarrays quantify 101-106 known RNA transcripts, and can be used to obtain an overview of gene expression patterns in muscle and fat (Wang et al., 2005; Tan et al., 2006). Genome-wide microarrays, which were used to profile gene expression in fat and muscle, are commercially available from Affymetrix and Agilent for bovine (Lee et al., 2010; Cassar-Malek et al., 2017; Hayashi et al., 2018; Liu et al., 2018), ovine (Liu et al., 2015; González-Calvo et al., 2017), porcine (Fontanesi et al., 2011; González-Prendes et al., 2017a,b) and chicken (Du et al., 2017; Zahoor et al., 2017; Chang et al., 2018) samples, however may be too expensive and provide an overwhelming volume of data for routine sample assessment. Additional commercial products for porcine samples were summarized by Pena et al. (2014), including microarrays specific for muscle. A customized bovine muscle/fat microarray with 9.6K probes was developed in 2005 (Lehnert et al., 2005). As with the genome-wide microarrays, it was mainly used to detect correlations between muscle traits and relevant biomarkers (Wang et al., 2005, 2009; Lehnert et al., 2007), but was also used to track transcriptomic changes during bovine adipogenesis in-vitro (Tan et al., 2006). However, this microarray was developed from a 2-year-old muscle and subcutaneous adipose tissue, and therefore, may miss specific genes expressed only in early developmental stages. RNA-seq is another genomic technique which quantifies the entire transcriptome de novo. Since this label-free technique requires minimal knowledge of the species at hand, it is advantageous for preliminary research of new exotic species or unknown RNA transcripts. Using this approach Sun et al. (2016) discovered a long non-coding RNA molecule that promotes myogenesis in bovine muscle by upregulating IGF-II expression. However RNA-seq is expensive, requires tedious analysis, and is less accurate for miRNA quantification (Horodyska et al., 2018), making it less pragmatic for clean meat research of extensively studied species (Reverter et al., 2006; Wickramasinghe et al., 2014).
miRNA, short RNA molecules that inhibit genes with complementary mRNA, play a crucial role in the regulation of muscle tissue development, including myogenesis (Wang et al., 2015; Dai et al., 2016; Zhang et al., 2017; Sadkowski et al., 2018) and adipogenesis (Yan et al., 2013). Several naturally occurring miRNA molecules have been shown to regulate myogenesis and adipogenesis in farm animals, including regulation of PPARG and C/EBPα expression (Romao et al., 2011) and lipid metabolism (Wilfred et al., 2007).
Protein expression can be measured using mass spectrometry. This can reveal novel biomarkers, such as tenderness and hypertrophy biomarkers (Picard et al., 2015), and provide a more reliable quantification of biological pathways (Poleti et al., 2018), especially regarding ECM deposition and post-translational modification (Chen et al., 2019), as compared to RNA measurements. Several reviews were published regarding proteomics and farm animals skeletal muscle research, discussing biomarkers and metabolic pathways, which are related to myogenesis, hypertrophy and meat traits, including flavor, tenderness, color, water holding capacity and pH (Picard et al., 2010, 2011; Bendixen et al., 2011; Veiseth-Kent et al., 2018).
Conclusion
Tissue engineering is a promising platform for creation of meat in a controlled, animal-free environment. It requires a shift in optimization parameters, with greater emphasis on inexpensive, animal-free biomaterials, efficient expansion of farm animal cells, tissue engineering of intramuscular fat, and analysis methods focusing on nutritional value and organoleptic properties. Implementation of tools and knowledge from the fields of food engineering and meat science will help develop novel research opportunities, such as tissue engineering of intramuscular fat or the generation of novel flavor-enhancing scaffolds. Clean meat scaffolds should be generated from edible biomaterials and designed to mimic the connective tissues of the muscle. Finally, multicellular seeding of muscle cells, fibroblasts, ECs and adipocytes inside such scaffolds can yield engineered muscle tissues with meat-like properties. Additional recommended reading materials can be found in Table 1.
Table 1
| Subject | Comments | Title | References |
|---|---|---|---|
| Tissue Engineering | Vascularized muscle tissues | Engineering vascularized skeletal muscle tissue | Levenberg et al., 2005 |
| Clean Meat | Tissue engineering | Principles of tissue engineering for food | Post and van der Weele, 2014 |
| Production and manufacturing | Opportunities for applying biomedical production and manufacturing methods to the development of the clean meat industry | Specht et al., 2018 | |
| Consumer acceptance | Consumer acceptance of cultured meat: A systematic review | Bryant and Barnett, 2018 | |
| Meat | Structure and composition | How muscle structure and composition influence meat and flesh quality | Listrat et al., 2016 |
| Flavor (book) | Flavor of meat and meat products | Shahidi, 2012 | |
| Developmental | Muscle development in farm animals | Fetal programming in meat production | Du et al., 2015 |
| Myogenic stem cells | SC—extended review | Satellite cells and the muscle stem cell niche | Yin et al., 2013 |
| SC protocols for different animals | Methods for animal satellite cell culture under a variety of conditions | Burton et al., 2000 | |
| Additional myogenic stem cells found in humans | Stem and progenitor Cells in skeletal muscle development, maintenance, and therapy | Péault et al., 2007 | |
| IMF | IMF in farm animals | Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers | Hocquette et al., 2010 |
| Adipogenesis protocols for clean meat | Adipogenesis from Bovine Precursors | Mehta et al., 2019 | |
| ECs | Vasculogenesis | Molecular regulation of vessel maturation | Jain, 2003 |
| Vascularization in tissue engineering | Vascularization—the conduit to viable engineered tissues | Kaully et al., 2009 | |
| Cell types and their potential in Vasculogenesis | Co-culture systems for Vasculogenesis | Perry et al., 2017a | |
| Angiocrine factors | Instructive role of the vascular niche in promoting tumor growth and tissue repair by angiocrine factors | Butler et al., 2010 | |
| ECM | Short review on fibrogenesis in farm-animals | Invited review: mesenchymal progenitor cells in intramuscular connective tissue development | Miao et al., 2016 |
| Longer review, focuses on humans | The importance of extracellular matrix in skeletal muscle development and function | Grzelkowska-Kowalczyk, 2016 | |
| Effect on meat quality | Intramuscular connective tissue and its role in meat quality | Purslow, 2005 | |
| Muscle derived fibroblasts | Skeletal muscle fibroblasts in health and disease | Chapman et al., 2016 | |
| Gene Expression | Transcriptomics and meat quality | Transcriptomics of meat quality | Guo and Dalrymple, 2017 |
| Biomaterials | Scaffolding | Biomaterials based strategies for skeletal muscle tissue engineering: existing technologies and future trends | Qazi et al., 2015 |
| Tissue Engineering: Focus on the Musculoskeletal System | Keeney et al., 2012 |
Additional reading materials.
Statements
Author contributions
TB-A conducted the literature survey, collated the relevant information, and wrote the paper with SL.
Funding
This work was supported by funding from Aleph-Farms Ltd Israel.
Acknowledgments
The authors wish to thank Alina Freiman for her graphical design assistance and Dr. Inbal Michael and Dr. Yehudit Posen for editorial assistance in preparing this manuscript.
Conflict of interest
SL is a co-founder and the CSO of Aleph Farms Ltd Israel. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
- AMR
Antimicrobial resistance
- BAM
Bio-artificial muscle
- BSC
Bovine satellite cells
- C/EBP
CCAAT/enhancer-binding protein
- EC
Endothelial cells
- ECM
Extracellular matrix
- ESC
Embryonic stem cells
- FABP4
Fatty acid binding protein 4
- FAP
Fibro/adipogenic precursors
- FAS
Fatty acid synthase
- FGF
Fibroblast growth factor
- G3P
Glycerol-3-phosphate
- GAG
Glycosaminoglycans
- GF
Growth factor
- GHG
Greenhouse gas
- HA
Hyaluronic acid
- hb-EGF
Heparin-binding EGF-like growth factor
- IGF-1
Insulin-like growth factor 1
- IMF
Intramuscular fat
- iPSC
Induced pluripotent stem cells
- LCFA
Long-chain fatty acids
- LIF
Leukemia inhibitory factor
- MSC
Mesenchymal stem cells
- PEG
Polyethylene glycol
- PG
Proteoglycan
- PLGA
Poly(lactic-co-glycolic acid)
- PLLA
Polylactic acid
- PPARG
Peroxisome proliferator-activated receptor γ
- SC
Satellite cells
- SVC
Stromal vascular cells
- TGF-β
Transforming growth factor beta.
Abbreviations
References
1
Aldridge A. Kouroupis D. Churchman S. English A. Ingham E. Jones E. (2013). Assay validation for the assessment of adipogenesis of multipotential stromal cells—a direct comparison of four different methods. Cytotherapy15, 89–101. 10.1016/j.jcyt.2012.07.001
2
Allen R. E. Boxhorn L. K. (1989). Regulation of skeletal muscle satellite cell proliferation and differentiation by transforming growth factor-beta, insulin-like growth factor I, and fibroblast growth factor. J. Cell. Physiol.138, 311–315. 10.1002/jcp.1041380213
3
Allison D. D. Grande-Allen K. J. (2006). Review. Hyaluronan: a powerful tissue engineering tool. Tissue Eng.12, 2131–2140. 10.1089/ten.2006.12.2131
4
Allt G. Lawrenson J. G. (2001). Pericytes: cell biology and pathology. Cells Tissues Organs169, 1–11. 10.1159/000047855
5
Anderson S. (2007). Determination of fat, moisture, and protein in meat and meat products by using the FOSS FoodScan near-infrared spectrophotometer with FOSS artificial neural network calibration model and associated database: collaborative study. J. AOAC Int.90, 1073–1083.
6
Anomaly J. (2015). What's wrong with factory farming?Public Health Ethics8, 246–254. 10.1093/phe/phu001
7
Austin L. Burgess A. W. (1991). Stimulation of myoblast proliferation in culture by leukaemia inhibitory factor and other cytokines. J. Neurol. Sci.101, 193–197. 10.1016/0022-510X(91)90045-9
8
Avesar J. Rosenfeld D. Truman-Rosentsvit M. Ben-Arye T. Geffen Y. Bercovici M. et al . (2017). Rapid phenotypic antimicrobial susceptibility testing using nanoliter arrays. Proc. Natl. Acad. Sci. U.S.A.114, E5787–E5795. 10.1073/pnas.1703736114
9
Aviss K. J. Gough J. E. Downes S. (2010). Aligned electrospun polymer fibres for skeletal muscle regeneration. Eur. Cell. Mater.19, 193–204. 10.22203/eCM.v019a19
10
Baum J. Duffy H. S. (2011). Fibroblasts and myofibroblasts: what are we talking about?J. Cardiovasc. Pharmacol.57, 376–379. 10.1097/FJC.0b013e3182116e39
11
Beier J. P. Klumpp D. Rudisile M. Dersch R. Wendorff J. H. Bleiziffer O. et al . (2009). Collagen matrices from sponge to nano: new perspectives for tissue engineering of skeletal muscle. BMC Biotechnology9:34. 10.1186/1472-6750-9-34
12
Bendixen E. Danielsen M. Hollung K. Gianazza E. Miller I. (2011). Farm animal proteomics–a review. J. Proteomics74, 282–293. 10.1016/j.jprot.2010.11.005
13
Bergers G. Song S. (2005). The role of pericytes in blood-vessel formation and maintenance. Neuro. Oncol.7, 452–464. 10.1215/S1152851705000232
14
Bernard C. Cassar-Malek I. Le Cunff M. Dubroeucq H. Renand G. Hocquette J.-F. (2007). New indicators of beef sensory quality revealed by expression of specific genes. J. Agric. Food Chem.55, 5229–5237. 10.1021/jf063372l
15
Bogliotti Y. S. Wu J. Vilarino M. Okamura D. Soto D. A. Zhong C. et al . (2018). Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc. Natl. Acad. Sci. U. S. A.115, 2090–2095. 10.1073/pnas.1716161115
16
Bonab M. M. Alimoghaddam K. Talebian F. Ghaffari S. H. Ghavamzadeh A. Nikbin B. (2006). Aging of mesenchymal stem cell in vitro. BMC Cell Biol.7:14. 10.1186/1471-2121-7-14
17
Bongiorni S. Gruber C. E. M. Bueno S. Chillemi G. Ferrè F. Failla S. et al . (2016). Transcriptomic investigation of meat tenderness in two Italian cattle breeds. Anim. Genet.47, 273–287. 10.1111/age.12418
18
Boonen K. J. M. Rosaria-Chak K. Y. Baaijens F. P. T. van der Schaft D. W. J. Post M. J. (2009). Essential environmental cues from the satellite cell niche: optimizing proliferation and differentiation. Am. J. Physiol. Cell Physiol.296, C1338–C1345. 10.1152/ajpcell.00015.2009
19
Bosnakovski D. Mizuno M. Kim G. Takagi S. Okumura M. Fujinaga T. (2005). Isolation and multilineage differentiation of bovine bone marrow mesenchymal stem cells. Cell Tissue Res.319, 243–253. 10.1007/s00441-004-1012-5
20
Brazile B. Lin S. Copeland K. M. Butler J. R. Cooley J. Brinkman-Ferguson E. et al . (2017). Chapter 6 - Ultrastructure and Biomechanics of Skeletal Muscle ECM: Implications in Tissue Regeneration, in Bio-Instructive Scaffolds for Musculoskeletal Tissue Engineering and Regenerative Medicine, eds BrownJ. L.KumbarS. G.BanikB. L. (Academic Press), 139–160. 10.1016/B978-0-12-803394-4.00006-9
21
Brett E. Chung N. Leavitt W. T. Momeni A. Longaker M. T. Wan D. C. (2017). A review of cell-based strategies for soft tissue reconstruction. Tissue Eng. Part B Rev.23, 336–346. 10.1089/ten.teb.2016.0455
22
Bridge K. Y. Smith C. K. Young R. B. (1998). Beta-adrenergic receptor gene expression in bovine skeletal muscle cells in culture. J. Anim. Sci.76, 2382–2391. 10.2527/1998.7692382x
23
Broad G. M. (2018). Effective animal advocacy: effective altruism, the social economy, and the animal protection movement. Agric. Human Values. 35, 777–789. 10.1007/s10460-018-9873-5
24
Broom D. M. (1991). Animal welfare: concepts and measurement. J. Anim. Sci.69, 4167–4175. 10.2527/1991.69104167x
25
Broom D. M. Fraser A. F. (2015). Domestic Animal Behaviour and Welfare, 5th Edn. CABI. Available online at: https://books.google.co.il/books?id=-uEcCgAAQBAJ (accessed September 13, 2018).
26
Bryant C. Barnett J. (2018). Consumer acceptance of cultured meat: a systematic review. Meat Sci.143, 8–17. 10.1016/j.meatsci.2018.04.008
27
Burton N. M. Vierck J. Krabbenhoft L. Bryne K. Dodson M. V. (2000). Methods for animal satellite cell culture under a variety of conditions. Methods Cell Sci.22, 51–61. 10.1023/A:1009830114804
28
Butler J. M. Kobayashi H. Rafii S. (2010). Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat. Rev. Cancer10, 138–146. 10.1038/nrc2791
29
Cai A. Horch R. E. Beier J. P. (2017). 15 - Nanofiber composites in skeletal muscle tissue engineering, in Nanofiber Composites for Biomedical Applications, eds RamalingamM.RamakrishnaS. (Woodhead Publishing), 369–394. 10.1016/B978-0-08-100173-8.00015-6
30
Calkins C. R. Hodgen J. M. (2007). A fresh look at meat flavor. Meat Sci.77, 63–80. 10.1016/j.meatsci.2007.04.016
31
Cao Y. (2007). Angiogenesis modulates adipogenesis and obesity. J. Clin. Invest.117, 2362–2368. 10.1172/JCI32239
32
Cao Y. (2013). Angiogenesis and vascular functions in modulation of obesity, adipose metabolism, and insulin sensitivity. Cell Metab.18, 478–489. 10.1016/j.cmet.2013.08.008
33
Cardoso T. C. Ferrari H. F. Garcia A. F. Novais J. B. Silva-Frade C. Ferrarezi M. C. et al . (2012). Isolation and characterization of Wharton's jelly-derived multipotent mesenchymal stromal cells obtained from bovine umbilical cord and maintained in a defined serum-free three-dimensional system. BMC Biotechnol.12:18. 10.1186/1472-6750-12-18
34
Cassar-Malek I. Boby C. Picard B. Reverter A. Hudson N. J. (2017). Molecular regulation of high muscle mass in developing Blonde d'Aquitaine cattle fetuses. Biol. Open6, 1483–1492. 10.1242/bio.024950
35
Chal J. Pourquié O. (2017). Making muscle: skeletal myogenesis in vivo and in vitro. Development144, 2104–2122. 10.1242/dev.151035
36
Chang K.-H. Liao H.-T. Chen J.-P. (2013). Preparation and characterization of gelatin/hyaluronic acid cryogels for adipose tissue engineering: in vitro and in vivo studies. Acta Biomater.9, 9012–9026. 10.1016/j.actbio.2013.06.046
37
Chang W. H. Zheng A. J. Chen Z. M. Zhang S. Cai H. Y. Liu G. H. (2018). β-Casomorphin increases fat deposition in broiler chickens by modulating expression of lipid metabolism genes. Animal13, 777–783. 10.1017/S1751731118002197
38
Chapman M. A. Meza R. Lieber R. L. (2016). Skeletal muscle fibroblasts in health and disease. Differentiation92, 108–115. 10.1016/j.diff.2016.05.007
39
Chaturvedi V. Dye D. E. Kinnear B. F. van Kuppevelt T. H. Grounds M. D. Coombe D. R. (2015). Interactions between skeletal muscle myoblasts and their extracellular matrix revealed by a serum free culture system. PLoS ONE10:e0127675. 10.1371/journal.pone.0127675
40
Chen D. Li W. Du M. Wu M. Cao B. (2015). Sequencing and characterization of divergent marbling levels in the beef cattle (Longissimus dorsi Muscle) transcriptome. Asian-australas. J. Anim. Sci.28, 158–165. 10.5713/ajas.14.0394
41
Chen F.-F. Wang Y.-Q. Tang G.-R. Liu S.-G. Cai R. Gao Y. et al . (2018). Differences between porcine longissimus thoracis and semitendinosus intramuscular fat content and the regulation of their preadipocytes during adipogenic differentiation. Meat Sci.147, 116–126. 10.1016/j.meatsci.2018.09.002
42
Chen L. Li Z. Everaert N. Lametsch R. Zhang D. (2019). Quantitative phosphoproteomic analysis of ovine muscle with different postmortem glycolytic rates. Food Chem.280, 203–209. 10.1016/j.foodchem.2018.12.056
43
Chen L. Todd R. Kiehlbauch J. Walters M. Kallen A. (2017). Notes from the field: pan-resistant new delhi metallo-beta-lactamase-producing klebsiella pneumoniae - washoe county, nevada, 2016. MMWR Morb. Mortal. Wkly. Rep.66:33. 10.15585/mmwr.mm6601a7
44
Chen Y. Bloemen V. Impens S. Moesen M. Luyten F. P. Schrooten J. (2011). Characterization and optimization of cell seeding in scaffolds by factorial design: quality by design approach for skeletal tissue engineering. Tissue Eng. Part C Methods17, 1211–1221. 10.1089/ten.tec.2011.0092
45
Chien K. B. Makridakis E. Shah R. N. (2013). Three-dimensional printing of soy protein scaffolds for tissue regeneration. Tissue Eng. Part C Methods19, 417–426. 10.1089/ten.tec.2012.0383
46
Choi J. S. Lee S. J. Christ G. J. Atala A. Yoo J. J. (2008). The influence of electrospun aligned poly(epsilon-caprolactone)/collagen nanofiber meshes on the formation of self-aligned skeletal muscle myotubes. Biomaterials29, 2899–2906. 10.1016/j.biomaterials.2008.03.031
47
Christov C. Chrétien F. Abou-Khalil R. Bassez G. Vallet G. Authier F.-J. et al . (2007). Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol. Biol. Cell18, 1397–1409. 10.1091/mbc.e06-08-0693
48
Ciccone J. (2015). Angioadaptive Allies: Examining the Relationship Between Endothelial Cells and Human Skeletal Muscle Myofibroblasts. Available online at: http://yorkspace.library.yorku.ca/xmlui/handle/10315/30133
49
Claeys E. De Smet S. Balcaen A. Raes K. Demeyer D. (2004). Quantification of fresh meat peptides by SDS–PAGE in relation to ageing time and taste intensity. Meat Sci.67, 281–288. 10.1016/j.meatsci.2003.11.001
50
Colwell A. S. Longaker M. T. Lorenz H. P. (2003). Fetal wound healing. Front. Biosci.8, s1240–8. 10.2741/1183
51
Conley A. J. Joensuu J. J. Menassa R. Brandle J. E. (2009). Induction of protein body formation in plant leaves by elastin-like polypeptide fusions. BMC Biol.7:48. 10.1186/1741-7007-7-48
52
Cosgrove B. D. Sacco A. Gilbert P. M. Blau H. M. (2009). A home away from home: challenges and opportunities in engineering in vitro muscle satellite cell niches. Differentiation78, 185–194. 10.1016/j.diff.2009.08.004
53
Council A. C. A. (2010). Contribution of the Pet Care Industry to the Australian Economy. East Hartford, CT: Rockwell Communications. Available online at: https://www.paws4play.com.au/pdfs/2010-Bis-Shrapnel-EconomPet-Benefits.pdf
54
Courtenay J. C. Sharma R. I. Scott J. L. (2018). Recent advances in modified cellulose for tissue culture applications. Molecules23:E654. 10.3390/molecules23030654
55
Cui H. X. Guo L. P. Zhao G. P. Liu R. R. Li Q. H. Zheng M. Q. et al . (2018). Method using a co-culture system with high-purity intramuscular preadipocytes and satellite cells from chicken pectoralis major muscle. Poult. Sci.97, 3691–3697. 10.3382/ps/pey023
56
Daamen W. F. van Moerkerk H. T. B. Hafmans T. Buttafoco L. Poot A. A. Veerkamp J. H. et al . (2003). Preparation and evaluation of molecularly-defined collagen–elastin–glycosaminoglycan scaffolds for tissue engineering. Biomaterials24, 4001–4009. 10.1016/S0142-9612(03)00273-4
57
Dai Y. Zhang W. R. Wang Y. M. Liu X. F. Li X. Ding X. B. et al . (2016). MicroRNA-128 regulates the proliferation and differentiation of bovine skeletal muscle satellite cells by repressing Sp1. Mol. Cell. Biochem.414, 37–46. 10.1007/s11010-016-2656-7
58
D'Alessandro A. Rinalducci S. Marrocco C. Zolla V. Napolitano F. Zolla L. (2012). Love me tender: an Omics window on the bovine meat tenderness network. J. Proteomics75, 4360–4380. 10.1016/j.jprot.2012.02.013
59
Damez J.-L. Clerjon S. (2013). Quantifying and predicting meat and meat products quality attributes using electromagnetic waves: an overview. Meat Sci.95, 879–896. 10.1016/j.meatsci.2013.04.037
60
Davidenko N. Campbell J. J. Thian E. S. Watson C. J. Cameron R. E. (2010). Collagen–hyaluronic acid scaffolds for adipose tissue engineering. Acta Biomater.6, 3957–3968. 10.1016/j.actbio.2010.05.005
61
Dawkins M. S. (2006). Through animal eyes: What behaviour tells us. Appl. Anim. Behav. Sci.100, 4–10. 10.1016/j.applanim.2006.04.010
62
De Deyne P. G. (2000). Formation of sarcomeres in developing myotubes: role of mechanical stretch and contractile activation. Am. J. Physiol. Cell Physiol.279, C1801–C1811. 10.1152/ajpcell.2000.279.6.C1801
63
Ding S. Swennen G. N. M. Messmer T. Gagliardi M. Molin D. G. M. Li C. et al . (2018). Maintaining bovine satellite cells stemness through p38 pathway. Sci. Rep.8:10808. 10.1038/s41598-018-28746-7
64
Discher D. E. Janmey P. Wang Y.-L. (2005). Tissue cells feel and respond to the stiffness of their substrate. Science310, 1139–1143. 10.1126/science.1116995
65
Dodson M. V. Allen R. E. Du M. Bergen W. G. Velleman S. G. Poulos S. P. et al . (2015). INVITED REVIEW: evolution of meat animal growth research during the past 50 years: adipose and muscle stem cells. J. Anim. Sci.93, 457–481. 10.2527/jas.2014-8221
66
Dodson M. V. Martin E. L. Brannon M. A. Mathison B. A. McFarland D. C. (1987). Optimization of bovine satellite cell-derived myotube formation in vitro. Tissue Cell19, 159–166.
67
Dodson M. V. Vierck J. L. Hossner K. L. Byrne K. McNamara J. P. (1997). The development and utility of a defined muscle and fat co-culture system. Tissue Cell29, 517–524.
68
Drury J. L. Mooney D. J. (2003). Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials24, 4337–4351. 10.1016/S0142-9612(03)00340-5
69
Du M. Huang Y. Das A. K. Yang Q. Duarte M. S. (2013). Manipulating mesenchymal progenitor cell differentiation to optimize performance and carcass value of beef cattle. J. Anim. Sci.91, 1419–1427. 10.2527/jas.2012-5670
70
Du M. Tong J. Zhao J. Underwood K. R. Zhu M. Ford S. P. et al . (2010a). Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci.88, E51–60. 10.2527/jas.2009-2311
71
Du M. Wang B. Fu X. Yang Q. Zhu M.-J. (2015). Fetal programming in meat production. Meat Sci.109, 40–47. 10.1016/j.meatsci.2015.04.010
72
Du M. Yin J. Zhu M. J. (2010b). Cellular signaling pathways regulating the initial stage of adipogenesis and marbling of skeletal muscle. Meat Sci.86, 103–109. 10.1016/j.meatsci.2010.04.027
73
Du Y. F. Ding Q. L. Li Y. M. Fang W. R. (2017). Identification of differentially expressed genes and pathways for myofiber characteristics in soleus muscles between chicken breeds differing in meat quality. Anim. Biotechnol.28, 83–93. 10.1080/10495398.2016.1206555
74
Duffy R. M. Sun Y. Feinberg A. W. (2016). Understanding the role of ecm protein composition and geometric micropatterning for engineering human skeletal muscle. Ann. Biomed. Eng.44, 2076–2089. 10.1007/s10439-016-1592-8
75
Duncan I. J. H. (2006). The changing concept of animal sentience. Appl. Anim. Behav. Sci.100, 11–19. 10.1016/j.applanim.2006.04.011
76
Edelman P. D. McFarland D. C. Mironov V. A. Matheny J. G. (2005). Commentary: In vitro-cultured meat production. Tissue Eng.11, 659–662. 10.1089/ten.2005.11.659
77
Egozi D. Shandalov Y. Freiman A. Rosenfeld D. Ben-Shimol D. Levenberg S. (2016). Engineered vascularized muscle flap. J. Vis. Exp. e52984. 10.3791/52984
78
Elder M. Fischer B. (2017). Focus on fish: a call to effective altruists. Essays Philos.18:7. 10.7710/1526-0569.1567
79
Elliott K. (2015). Antibiotics on the Farm: Agriculture's Role in Drug Resistance. CGD Policy Paper 59. Available online at: https://www.cgdev.org/sites/default/files/CGD-Policy-Paper-59-Elliott-Antibiotics-Farm-Agriculture-Drug-Resistance.pdf
80
Engler A. J. Sen S. Sweeney H. L. Discher D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell126, 677–689. 10.1016/j.cell.2006.06.044
81
Enrione J. Díaz-Calderón P. Weinstein-Oppenheimer C. R. Sánchez E. Fuentes M. A. Brown D. I. et al . (2013). Designing a gelatin/chitosan/hyaluronic acid biopolymer using a thermophysical approach for use in tissue engineering. Bioprocess Biosyst. Eng.36, 1947–1956. 10.1007/s00449-013-0971-x
82
Eom J. Feisst V. Ranjard L. Loomes K. Damani T. Jackson-Patel V. et al . (2018). Visualization and quantification of mesenchymal cell adipogenic differentiation potential with a lineage specific marker. J. Vis. Exp.133:e57153. 10.3791/57153
83
Eshel G. Shepon A. Makov T. Milo R. (2014). Land, irrigation water, greenhouse gas, and reactive nitrogen burdens of meat, eggs, and dairy production in the United States. Proc. Natl. Acad. Sci. U. S. A.111, 11996–12001. 10.1073/pnas.1402183111
84
Fedorovich N. E. Alblas J. de Wijn J. R. Hennink W. E. Verbout A. J. Dhert W. J. A. (2007). Hydrogels as extracellular matrices for skeletal tissue engineering: state-of-the-art and novel application in organ printing. Tissue Eng.13, 1905–1925. 10.1089/ten.2006.0175
85
Flachowsky G. Meyer U. Südekum K.-H. (2018). Invited review: Resource inputs and land, water and carbon footprints from the production of edible protein of animal origin. Arch. Anim. Breed.61, 17–36. 10.5194/aab-61-17-2018
86
Fontanesi L. Galimberti G. Calò D. G. Colombo M. Astolfi A. Formica S. et al . (2011). Microarray gene expression analysis of porcine skeletal muscle sampled at several post mortem time points. Meat Sci.88, 604–609. 10.1016/j.meatsci.2011.02.001
87
Forbes S. J. Rosenthal N. (2014). Preparing the ground for tissue regeneration: from mechanism to therapy. Nat. Med.20, 857–869. 10.1038/nm.3653
88
Freiman A. Shandalov Y. Rosenfeld D. Shor E. Ben-David D. Meretzki S. et al . (2018). Engineering vascularized flaps using adipose-derived microvascular endothelial cells and mesenchymal stem cells. J. Tissue Eng. Regen. Med.12, e130–e141. 10.1002/term.2436
89
Frey R. S. Johnson B. J. Hathaway M. R. White M. E. Dayton W. R. (1995). Growth factor responsiveness of primary satellite cell cultures from steers implanted with trenbolone acetate and estradiol-17β. Basic Appl. Myol.5, 71–79.
90
Friedman N. D. Temkin E. Carmeli Y. (2016). The negative impact of antibiotic resistance. Clin. Microbiol. Infect.22, 416–422. 10.1016/j.cmi.2015.12.002
91
Fu S. Zhao Y. Li Y. Li G. Chen Y. Li Z. et al . (2018). Characterization of miRNA transcriptome profiles related to breast muscle development and intramuscular fat deposition in chickens. J. Cell. Biochem.119, 7063–7079. 10.1002/jcb.27024
92
Fuoco C. Petrilli L. L. Cannata S. Gargioli C. (2016). Matrix scaffolding for stem cell guidance toward skeletal muscle tissue engineering. J. Orthop. Surg. Res.11:86. 10.1186/s13018-016-0421-y
93
Gasteratos K. S. Sherman R. (2018). Consumer Interest Towards Clean Meat. Available online at: https://dash.harvard.edu/handle/1/34901168
94
Gaydhane M. K. Mahanta U. Sharma C. S. Khandelwal M. Ramakrishna S. (2018). Cultured meat: state of the art and future. Biomanufact. Rev.3:1. 10.1007/s40898-018-0005-1
95
Ge Y. Sun Y. Chen J. (2011). IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol.192, 69–81. 10.1083/jcb.201007165
96
Gerber P. J. Steinfeld H. Henderson B. Mottet A. Opio C. Dijkman J. et al . (2013). Tackling Climate Change Through Livestock: A Global Assessment of Emissions and Mitigation Opportunities.Rome: Food and Agriculture Organization of the United Nations (FAO).
97
Ghasemi-Mobarakeh L. Prabhakaran M. P. Tian L. Shamirzaei-Jeshvaghani E. Dehghani L. Ramakrishna S. (2015). Structural properties of scaffolds: Crucial parameters towards stem cells differentiation. World J. Stem Cells7, 728–744. 10.4252/wjsc.v7.i4.728
98
Gholobova D. Decroix L. Van Muylder V. Desender L. Gerard M. Carpentier G. et al . (2015). Endothelial network formation within human tissue-engineered skeletal muscle. Tissue Eng. Part A21, 2548–2558. 10.1089/ten.tea.2015.0093
99
Gilbert T. W. Stolz D. B. Biancaniello F. Simmons-Byrd A. Badylak S. F. (2005). Production and characterization of ECM powder: implications for tissue engineering applications. Biomaterials26, 1431–1435. 10.1016/j.biomaterials.2004.04.042
100
Gilbert-López B. Valdés A. Acunha T. García-Cañas V. Simó C. Cifuentes A. (2017). Foodomics: LC and LC-MS-based omics strategies in food science and nutrition, in Liquid Chromatography, 2nd Edn, eds FanaliS.HaddadP. R.PooleC. F.RiekkolaM-L. (Amsterdam: Elsevier), 267–299. 10.1016/B978-0-12-805392-8.00010-4
101
Gillies A. R. Lieber R. L. (2011). Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve44, 318–331. 10.1002/mus.22094
102
Gomillion C. T. Burg K. J. L. (2006). Stem cells and adipose tissue engineering. Biomaterials27, 6052–6063. 10.1016/j.biomaterials.2006.07.033
103
González-Calvo L. Dervishi E. Joy M. Sarto P. Martin-Hernandez R. Serrano M. et al . (2017). Genome-wide expression profiling in muscle and subcutaneous fat of lambs in response to the intake of concentrate supplemented with vitamin E. BMC Genomics18:92. 10.1186/s12864-016-3405-8
104
González-Prendes R. Quintanilla R. Amills M. (2017a). Investigating the genetic regulation of the expression of 63 lipid metabolism genes in the pig skeletal muscle. Anim. Genet.48, 606–610. 10.1111/age.12586
105
González-Prendes R. Quintanilla R. Cánovas A. Manunza A. Figueiredo Cardoso T. Jordana J. et al . (2017b). Joint QTL mapping and gene expression analysis identify positional candidate genes influencing pork quality traits. Sci. Rep.7:39830. 10.1038/srep39830
106
Goodwin J. N. Shoulders C. W. (2013). The future of meat: a qualitative analysis of cultured meat media coverage. Meat Sci.95, 445–450. 10.1016/j.meatsci.2013.05.027
107
Gorraiz C. Beriain M. J. Chasco J. Insausti K. (2002). Effect of aging time on volatile compounds, odor, and flavor of cooked beef from pirenaica and friesian bulls and heifers. J. Food Sci.67, 916–922. 10.1111/j.1365-2621.2002.tb09428.x
108
Gottipamula S. Muttigi M. S. Chaansa S. Ashwin K. M. Priya N. Kolkundkar U. et al . (2016). Large-scale expansion of pre-isolated bone marrow mesenchymal stromal cells in serum-free conditions. J. Tissue Eng. Regen. Med.10, 108–119. 10.1002/term.1713
109
Grahl S. Palanisamy M. Strack M. Meier-Dinkel L. Toepfl S. Mörlein D. (2018). Towards more sustainable meat alternatives: How technical parameters affect the sensory properties of extrusion products derived from soy and algae. J. Clean. Prod.198, 962–971. 10.1016/j.jclepro.2018.07.041
110
Grant A. C. Ortiz-Colòn G. Doumit M. E. Buskirk D. D. (2008). Optimization of in vitro conditions for bovine subcutaneous and intramuscular preadipocyte differentiation. J. Anim. Sci.86, 73–82. 10.2527/jas.2007-0379
111
Green C. (2008). How to End 98% of Animal Abuse in the Next 25 Years. Available online at: https://faunalytics.org/how-to-end-98-of-animal-abuse-in-the-next-25-years (accessed September 13, 2018).
112
Greger M. (2007). The human/animal interface: emergence and resurgence of zoonotic infectious diseases. Crit. Rev. Microbiol.33, 243–299. 10.1080/10408410701647594
113
Griffith C. K. Miller C. Sainson R. C. A. Calvert J. W. Jeon N. L. Hughes C. C. W. et al . (2005). Diffusion limits of an in vitro thick prevascularized tissue. Tissue Eng.11, 257–266. 10.1089/ten.2005.11.257
114
Grzelkowska-Kowalczyk K. (2016). The importance of extracellular matrix in skeletal muscle development and function, in Composition and Function of the Extracellular Matrix in the Human Body, ed. F. Travascio (InTech), 3–24. 10.5772/62230
115
Guan L. Hu X. Liu L. Xing Y. Zhou Z. Liang X. et al . (2017). bta-miR-23a involves in adipogenesis of progenitor cells derived from fetal bovine skeletal muscle. Sci. Rep.7:43716. 10.1038/srep43716
116
Guo B. Dalrymple B. P. (2017). Chapter 11 - Transcriptomics of Meat Quality, in New Aspects of Meat Quality, ed. P. P. Purslow (Woodhead Publishing), 259–320. 10.1016/B978-0-08-100593-4.00012-6
117
Guo B. Greenwood P. L. Cafe L. M. Zhou G. Zhang W. Dalrymple B. P. (2015). Transcriptome analysis of cattle muscle identifies potential markers for skeletal muscle growth rate and major cell types. BMC Genomics16:177. 10.1186/s12864-015-1403-x
118
Halbleib M. Skurk T. de Luca C. von Heimburg D. Hauner H. (2003). Tissue engineering of white adipose tissue using hyaluronic acid-based scaffolds. I: in vitro differentiation of human adipocyte precursor cells on scaffolds. Biomaterials24, 3125–3132. 10.1016/S0142-9612(03)00156-X
119
Harper G. S. Pethick D. W. (2004). How might marbling begin?Aust. J. Exp. Agric.44, 653–662. 10.1071/EA02114
120
Harrysson A. Lövgren A. (2012). Method for Production of Recombinant Human Thrombin. US Patent. Available online at: https://patentimages.storage.googleapis.com/5a/9b/7e/0754ac5cda8493/US8206967.pdf (accessed October 2, 2018).
121
Hausman G. J. (2012). Meat Science and Muscle Biology Symposium: the influence of extracellular matrix on intramuscular and extramuscular adipogenesis. J. Anim. Sci.90, 942–949. 10.2527/jas.2011-4616
122
Hausman G. J. Poulos S. (2004). Recruitment and differentiation of intramuscular preadipocytes in stromal-vascular cell cultures derived from neonatal pig semitendinosus muscles. J. Anim. Sci.82, 429–437. 10.2527/2004.822429x
123
Hayashi M. Kido K. Hodate K. (2018). Microarray analysis of Longissimus thoracis muscle gene expressions in vitamin A-restricted Japanese Black steers in middle fattening stage. Anim. Sci. J.89, 88–96. 10.1111/asj.12898
124
Heher P. Maleiner B. Prüller J. Teuschl A. H. Kollmitzer J. Monforte X. et al . (2015). A novel bioreactor for the generation of highly aligned 3D skeletal muscle-like constructs through orientation of fibrin via application of static strain. Acta Biomater.24, 251–265. 10.1016/j.actbio.2015.06.033
125
Hemmrich K. von Heimburg D. Rendchen R. Di Bartolo C. Milella E. Pallua N. (2005). Implantation of preadipocyte-loaded hyaluronic acid-based scaffolds into nude mice to evaluate potential for soft tissue engineering. Biomaterials26, 7025–7037. 10.1016/j.biomaterials.2005.04.065
126
Herzog H. (2016). Farm Animal Suffering Leaves a Bad Taste In Your Mouth. Psychol. Today. Available onlien at: https://works.bepress.com/harold-herzog/98/download/ (accessed October 03, 2018).
127
Hickey R. J. Modulevsky D. J. Cuerrier C. M. Pelling A. E. (2018). Customizing the shape and microenvironment biochemistry of biocompatible macroscopic plant-derived cellulose scaffolds. ACS Biomater. Sci. Eng.11, 3726–3736. 10.1021/acsbiomaterials.8b00178
128
Hirashima M. Imamura T. Yano K. Kawamura R. Meta A. Tokieda Y. et al . (2016). High-level expression and preparation of recombinant human fibrinogen as biopharmaceuticals. J. Biochem.159, 261–270. 10.1093/jb/mvv099
129
Hocquette J.-F. Lehnert S. Barendse W. Cassar-Malek I. Picard B. (2007). Recent advances in cattle functional genomics and their application to beef quality. Animal1, 159–173. 10.1017/S1751731107658042
130
Hocquette J. F. Gondret F. Baéza E. Médale F. Jurie C. Pethick D. W. (2010). Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Animal4, 303–319. 10.1017/S1751731109991091
131
Hong L. Peptan I. Clark P. Mao J. J. (2005). Ex vivo adipose tissue engineering by human marrow stromal cell seeded gelatin sponge. Ann. Biomed. Eng.33, 511–517. 10.1007/s10439-005-2510-7
132
Hopkins P. D. Dacey A. (2008). Vegetarian meat: could technology save animals and satisfy meat eaters?J. Agric. Environ. Ethics21, 579–596. 10.1007/s10806-008-9110-0
133
Horodyska J. Oster M. Reyer H. Mullen A. M. Lawlor P. G. Wimmers K. et al . (2018). Analysis of meat quality traits and gene expression profiling of pigs divergent in residual feed intake. Meat Sci.137, 265–274. 10.1016/j.meatsci.2017.11.021
134
Huang G. Li F. Zhao X. Ma Y. Li Y. Lin M. et al . (2017). Functional and biomimetic materials for engineering of the three-dimensional cell microenvironment. Chem. Rev.117, 12764–12850. 10.1021/acs.chemrev.7b00094
135
Huang J. Huang K. You X. Liu G. Hollett G. Kang Y. et al . (2018). Evaluation of tofu as a potential tissue engineering scaffold. J. Mater. Chem. B Mater. Biol. Med.6, 1328–1334. 10.1039/C7TB02852K
136
Huang Y. Das A. K. Yang Q.-Y. Zhu M.-J. Du M. (2012). Zfp423 promotes adipogenic differentiation of bovine stromal vascular cells. PLoS ONE7:e47496. 10.1371/journal.pone.0047496
137
Hudson N. J. Reverter A. Greenwood P. L. Guo B. Cafe L. M. Dalrymple B. P. (2015). Longitudinal muscle gene expression patterns associated with differential intramuscular fat in cattle. Animal9, 650–659. 10.1017/S1751731114002754
138
Hughes D. (1995). Animal welfare: the consumer and the food industry. Br. Food J.97, 3–7. 10.1108/00070709510104529
139
Jaffe E. A. (2012). Biology of Endothelial Cells. Available online at: https://books.google.co.in/books?id=Y6jSBwAAQBAJ&redir_esc=y (accessed September 11, 2018).
140
Jain R. K. (2003). Molecular regulation of vessel maturation. Nat. Med.9, 685–693. 10.1038/nm0603-685
141
Jain R. K. Au P. Tam J. Duda D. G. Fukumura D. (2005). Engineering vascularized tissue. Nat. Biotechnol.23, 821–823. 10.1038/nbt0705-821
142
Jana S. Cooper A. Zhang M. (2013). Chitosan scaffolds with unidirectional microtubular pores for large skeletal myotube generation. Adv. Healthc. Mater. 2, 557–561. 10.1002/adhm.201200177
143
Jana S. Levengood S. K. L. Zhang M. (2016). Anisotropic materials for skeletal-muscle-tissue engineering. Adv. Mater.28, 10588–10612. 10.1002/adma.201600240
144
Jockenhoevel S. Zund G. Hoerstrup S. P. Chalabi K. Sachweh J. S. Demircan L. et al . (2001). Fibrin gel–advantages of a new scaffold in cardiovascular tissue engineering. Eur. J. Cardiothorac. Surg.19, 424–430. 10.1016/S1010-7940(01)00624-8
145
Jordan G. Thomasius R. Schröder H. Wulf J. S. Schlüter O. Sumpf B. et al . (2009). Non-invasive mobile monitoring of meat quality. J. für Verbrauchers. Lebensmittelsicher.4, 7–14. 10.1007/s00003-009-0389-1
146
Kadler K. E. Baldock C. Bella J. Boot-Handford R. P. (2007). Collagens at a glance. J. Cell Sci.120, 1955–1958. 10.1242/jcs.03453
147
Kamanga-Sollo E. Pampusch M. S. Xi G. White M. E. Hathaway M. R. Dayton W. R. (2004). IGF-I mRNA levels in bovine satellite cell cultures: effects of fusion and anabolic steroid treatment. J. Cell. Physiol.201, 181–189. 10.1002/jcp.20000
148
Kamanga-Sollo E. White M. E. Hathaway M. R. Chung K. Y. Johnson B. J. Dayton W. R. (2008). Roles of IGF-I and the estrogen, androgen and IGF-I receptors in estradiol-17β- and trenbolone acetate-stimulated proliferation of cultured bovine satellite cells. Domest. Anim. Endocrinol.35, 88–97. 10.1016/j.domaniend.2008.02.003
149
Kang J. H. Gimble J. M. Kaplan D. L. (2009). In vitro 3D model for human vascularized adipose tissue. Tissue Eng. Part A15, 2227–2236. 10.1089/ten.tea.2008.0469
150
Kaully T. Kaufman-Francis K. Lesman A. Levenberg S. (2009). Vascularization—the conduit to viable engineered tissues. Tissue Eng. Part B Rev.15, 159–169. 10.1089/ten.teb.2008.0193
151
Keeney M. Han L.-H. Onyiah S. Yang F. (2012). Tissue Engineering: Focus on the Musculoskeletal System. Biomaterials Science: An Integrated Clinical and Engineering Approach, 191. Available online at: https://books.google.co.in/books?id=kcoeYDazWgIC&redir_esc=y (accessed October 02, 2018).
152
King J. A. Miller W. M. (2007). Bioreactor development for stem cell expansion and controlled differentiation. Curr. Opin. Chem. Biol.11, 394–398. 10.1016/j.cbpa.2007.05.034
153
Kjaer M. (2004). Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol. Rev.84, 649–698. 10.1152/physrev.00031.2003
154
Koffler J. Kaufman-Francis K. Shandalov Y. Yulia S. Egozi D. Dana E. et al . (2011). Improved vascular organization enhances functional integration of engineered skeletal muscle grafts. Proc. Natl. Acad. Sci. U. S. A.108, 14789–14794. 10.1073/pnas.1017825108
155
Kollias H. D. McDermott J. C. (2008). Transforming growth factor-β and myostatin signaling in skeletal muscle. J. Appl. Physiol.104, 579–587. 10.1152/japplphysiol.01091.2007
156
Koning M. Harmsen M. C. van Luyn M. J. A. Werker P. M. N. (2009). Current opportunities and challenges in skeletal muscle tissue engineering. J. Tissue Eng. Regen. Med.3, 407–415. 10.1002/term.190
157
Koohmaraie M. Kent M. P. Shackelford S. D. Veiseth E. Wheeler T. L. (2002). Meat tenderness and muscle growth: is there any relationship?Meat Sci.62, 345–352. 10.1016/S0309-1740(02)00127-4
158
Kragstrup T. W. Kjaer M. Mackey A. L. (2011). Structural, biochemical, cellular, and functional changes in skeletal muscle extracellular matrix with aging. Scand. J. Med. Sci. Sports21, 749–757. 10.1111/j.1600-0838.2011.01377.x
159
Kramer A. Szczesniak A. S. (eds). (1973). Texture Measurements of Foods: Psychophysical Fundamentals: Sensory, Mechanical, and Chemical Procedures, and Their Interrelationships. Dordrecht: Springer.
160
Kraus N. A. Ehebauer F. Zapp B. Rudolphi B. Kraus B. J. Kraus D. (2016). Quantitative assessment of adipocyte differentiation in cell culture. Adipocyte5, 351–358. 10.1080/21623945.2016.1240137
161
Krieger J. Park B.-W. Lambert C. R. Malcuit C. (2018). 3D skeletal muscle fascicle engineering is improved with TGF-β1 treatment of myogenic cells and their co-culture with myofibroblasts. PeerJ6:e4939. 10.7717/peerj.4939
162
Kümmerer K. (2003). Significance of antibiotics in the environment. J. Antimicrob. Chemother. 52, 5–7. 10.1093/jac/dkg293
163
Kunz-Schughart L. A. Schroeder J. A. Wondrak M. van Rey F. Lehle K. Hofstaedter F. et al . (2006). Potential of fibroblasts to regulate the formation of three-dimensional vessel-like structures from endothelial cells in vitro. Am. J. Physiol. Cell Physiol.290, C1385–C1398. 10.1152/ajpcell.00248.2005
164
Laliotis G. P. Bizelis I. Rogdakis E. (2010). Comparative approach of the de novo fatty acid synthesis (Lipogenesis) between ruminant and non ruminant mammalian species: from biochemical level to the main regulatory lipogenic genes. Curr. Genomics11, 168–183. 10.2174/138920210791110960
165
Landau S. Guo S. Levenberg S. (2018). Localization of engineered vasculature within 3D tissue constructs. Front Bioeng Biotechnol6:2. 10.3389/fbioe.2018.00002
166
Landau S. Szklanny A. A. Yeo G. C. Shandalov Y. Kosobrodova E. Weiss A. S. et al . (2017). Tropoelastin coated PLLA-PLGA scaffolds promote vascular network formation. Biomaterials122, 72–82. 10.1016/j.biomaterials.2017.01.015
167
Langelaan M. L. P. Boonen K. J. M. Polak R. B. Baaijens F. P. T. Post M. J. van der Schaft D. W. J. (2010). Meet the new meat: tissue engineered skeletal muscle. Trends Food Sci. Technol.21, 59–66. 10.1016/j.tifs.2009.11.001
168
Langelaan M. L. P. Boonen K. J. M. Rosaria-Chak K. Y. van der Schaft D. W. J. Post M. J. Baaijens F. P. T. (2011). Advanced maturation by electrical stimulation: differences in response between C2C12 and primary muscle progenitor cells. J. Tissue Eng. Regen. Med.5, 529–539. 10.1002/term.345
169
Lanza R. Langer R. Vacanti J. P. (2011). Principles of Tissue Engineering. Academic Press. Available online at: https://market.android.com/details?id=book-rAFkm_4hOIMC (accessed September 02, 2018).
170
Lapin M. R. Gonzalez J. M. Johnson S. E. (2013). Substrate elasticity affects bovine satellite cell activation kinetics in vitro. J. Anim. Sci.91, 2083–2090. 10.2527/jas.2012-5732
171
Lee E. J. Malik A. Pokharel S. Ahmad S. Mir B. A. Cho K. H. et al . (2014). Identification of genes differentially expressed in myogenin knock-down bovine muscle satellite cells during differentiation through RNA sequencing analysis. PLoS ONE9:e92447. 10.1371/journal.pone.0092447
172
Lee S.-H. Gondro C. van der Werf J. Kim N.-K. Lim D.-J. Park E.-W. et al . (2010). Use of a bovine genome array to identify new biological pathways for beef marbling in Hanwoo (Korean Cattle). BMC Genomics11:623. 10.1186/1471-2164-11-623
173
Lehnert S. A. Reverter A. Byrne K. A. Wang Y. Nattrass G. S. Hudson N. J. et al . (2007). Gene expression studies of developing bovine longissimus muscle from two different beef cattle breeds. BMC Dev. Biol.7:95. 10.1186/1471-213X-7-95
174
Lehnert S. A. Wang Y. H. Byrne K. A. (2005). Development and application of a bovine cDNA microarray for expression profiling of muscle and adipose tissue. Aust. J. Exp. Agric.44, 1127–1133. 10.1071/EA03238
175
Lengi A. J. Corl B. A. (2010). Factors influencing the differentiation of bovine preadipocytes in vitro. J. Anim. Sci.88, 1999–2008. 10.2527/jas.2009-2439
176
Lesman A. Koffler J. Atlas R. Blinder Y. J. Kam Z. (2011). Engineering vessel-like networks within multicellular fibrin-based constructs. Biomaterials.32, 7856–7869. 10.1016/j.biomaterials.2011.07.003
177
Levenberg S. Rouwkema J. Macdonald M. Garfein E. S. Kohane D. S. Darland D. C. et al . (2005). Engineering vascularized skeletal muscle tissue. Nat. Biotechnol.23, 879–884. 10.1038/nbt1109
178
Levy-Mishali M. Zoldan J. Levenberg S. (2009). Effect of scaffold stiffness on myoblast differentiation. Tissue Eng. Part A15, 935–944. 10.1089/ten.tea.2008.0111
179
Linde D. A. (2018). Gene Expression Profiles in Longissimus Dorsi Muscle of Bonsmara and Nguni Cattle Fed Two Different Diets. Available online at: https://repository.up.ac.za/handle/2263/65906
180
Listrat A. Lebret B. Louveau I. Astruc T. Bonnet M. Lefaucheur L. et al . (2016). How muscle structure and composition influence meat and flesh quality. Sci. World J.2016:3182746. 10.1155/2016/3182746
181
Liu G. Liu X. Shahzad K. You W. Loor J. J. Wan F. (2018). Bioinformatics analyses of bovine adipose tissue transcriptome from lilu beef cattle at different stages of growth. PJZ50:1855. 10.17582/journal.pjz/2018.50.5.1847.1855
182
Liu N. He J. N. Yu W. M. Liu K. D. Cheng M. Liu J. F. et al . (2015). Transcriptome analysis of skeletal muscle at prenatal stages in polled dorset versus small-tailed han sheep. Genet. Mol. Res.14, 1085–1095. 10.4238/2015.February.6.12
183
Loh Q. L. Choong C. (2013). Three-dimensional scaffolds for tissue engineering applications: role of porosity and pore size. Tissue Eng. Part B Rev.19, 485–502. 10.1089/ten.teb.2012.0437
184
Lovett M. Lee K. Edwards A. Kaplan D. L. (2009). Vascularization strategies for tissue engineering. Tissue Eng. Part B Rev.15, 353–370. 10.1089/ten.teb.2009.0085
185
Luckett C. R. Kuttappan V. A. Johnson L. G. Owens C. M. Seo H.-S. (2014). Comparison of three instrumental methods for predicting sensory texture attributes of poultry deli meat: texture of poultry deli meat. J. Sens. Stud.29, 171–181. 10.1111/joss.12092
186
Lusk J. L. Norwood F. B. (2011). Animal welfare economics. Applied Economic Perspectives. Available online at: https://academic.oup.com/aepp/article-abstract/33/4/463/8220
187
Ma Y. N. Wang B. Wang Z. X. Gomez N. A. Zhu M. J. Du M. (2018). Three-dimensional spheroid culture of adipose stromal vascular cells for studying adipogenesis in beef cattle. Animal12, 1–7. 10.1017/S1751731118000150
188
Mackey A. L. Magnan M. Chazaud B. Kjaer M. (2017). Human skeletal muscle fibroblasts stimulate in vitro myogenesis and in vivo muscle regeneration. J. Physiol.595, 5115–5127. 10.1113/JP273997
189
Meadus W. Duff P. Juarez M. Roberts J. Zantinge J. (2018). Identification of marbling gene loci in commercial pigs in canadian herds. Collect. FAO Agric.8:122. 10.3390/agriculture8080122
190
Mehta F. Theunissen R. Post M. J. (2019). Adipogenesis from Bovine Precursors, in Myogenesis: Methods and Protocols, ed. S. B. Rønning (New York, NY: Springer), 111–125. 10.1007/978-1-4939-8897-6_8
191
Mesa Ospina N. Ospina Alvarez S. P. Escobar Sierra D. M. Rojas Vahos D. F. Zapata Ocampo P. A. Ossa Orozco C. P. (2015). Isolation of chitosan from Ganoderma lucidum mushroom for biomedical applications. J. Mater. Sci. Mater. Med.26:135. 10.1007/s10856-015-5461-z
192
Miao Z. G. Zhang L. P. Fu X. Yang Q. Y. Zhu M. J. Dodson M. V. et al . (2016). Invited review: mesenchymal progenitor cells in intramuscular connective tissue development. Animal10, 75–81. 10.1017/S1751731115001834
193
Miller M. F. Carr M. A. Ramsey C. B. Crockett K. L. Hoover L. C. (2001). Consumer thresholds for establishing the value of beef tenderness. J. Anim. Sci.79, 3062–3068. 10.2527/2001.79123062x
194
Mitchell A. D. (2007). Impact of research with cattle, pigs, and sheep on nutritional concepts: body composition and growth. J. Nutr.137, 711–714. 10.1093/jn/137.3.711
195
Monte D. F. Mem A. Fernandes M. R. Cerdeira L. Esposito F. Galvão J. A. et al . (2017). Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother.61:e02718–16. 10.1128/AAC.02718-16
196
Moritz M. S. M. Verbruggen S. E. L. Post M. J. (2015). Alternatives for large-scale production of cultured beef: A review. J. Integr. Agric.14, 208–216. 10.1016/S2095-3119(14)60889-3
197
Moseti D. Regassa A. Kim W.-K. (2016). Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. Int. J. Mol. Sci.17:124. 10.3390/ijms17010124
198
Muroya S. Nakajima I. Chikuni K. (2001). Bovine skeletal muscle cells predominantly express a vascular cell adhesion molecule-1 seven-ig domain splice form. Zoolog. Sci.18, 797–805. 10.2108/zsj.18.797
199
Nekanti U. Rao V. B. Bahirvani A. G. Jan M. Totey S. Ta M. (2010). Long-term expansion and pluripotent marker array analysis of Wharton's jelly-derived mesenchymal stem cells. Stem Cells Dev.19, 117–130. 10.1089/scd.2009.0177
200
Neubauer M. Hacker M. Bauer-Kreisel P. (2005). Adipose tissue engineering based on mesenchymal stem cells and basic fibroblast growth factor in Vitro. Tissue11:1840–1851. 10.1089/ten.2005.11.1840
201
Nguyen N. T. Nguyen H. M. Nguyen C. V. Nguyen T. V. Nguyen M. T. Thai H. Q. et al . (2016). Use of colistin and other critical antimicrobials on pig and chicken farms in southern vietnam and its association with resistance in commensal escherichia coli bacteria. Appl. Environ. Microbiol.82, 3727–3735. 10.1128/AEM.00337-16
202
Nicola N. A. Babon J. J. (2015). Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev.26, 533–544. 10.1016/j.cytogfr.2015.07.001
203
Nollet L. M. L. (ed.). (2012). Handbook of Meat, Poultry and Seafood Quality.Oxford, UK: Blackwell Publishing Ltd. 10.1002/9781118352434
204
Oerther S. Le Gall H. Payan E. Lapicque F. Presle N. Hubert P. et al . (1999). Hyaluronate-alginate gel as a novel biomaterial: Mechanical properties and formation mechanism. Biotechnol. Bioeng.63, 206–215.
205
Oguz S. Tam N. Aydogdu A. Sumnu G. Sahin S. (2018). Development of novel pea flour-based nanofibres by electrospinning method. Int. J. Food Sci. Technol.53, 1269–1277. 10.1111/ijfs.13707
206
Okamura L. H. Cordero P. Palomino J. Parraguez V. H. Torres C. G. Peralta O. A. (2018). Myogenic differentiation potential of mesenchymal stem cells derived from fetal bovine bone marrow. Anim. Biotechnol.29, 1–11. 10.1080/10495398.2016.1276926
207
O'Neill J. (2014). Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist20, 1–16.
208
Oswald J. Boxberger S. Jørgensen B. Feldmann S. Ehninger G. Bornhäuser M. et al . (2004). Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells22, 377–384. 10.1634/stemcells.22-3-377
209
Pan Y. Jing J. Qiao L. Liu J. Zhao J. An L. et al . (2018). miR-124-3p affects the formation of intramuscular fat through alterations in branched chain amino acid consumption in sheep. Biochem. Biophys. Res. Commun.495, 1769–1774. 10.1016/j.bbrc.2017.12.046
210
Pandurangan M. Kim D. H. (2015). A novel approach for in vitro meat production. Appl. Microbiol. Biotechnol.99, 5391–5395. 10.1007/s00253-015-6671-5
211
Panskeep J. Others (2012). Low P. Cambridge declaration of consciousness, in Frances Crick Memorial Conference on Consciousness in Human and non-Human Animals at Churchill College, University of Cambridge (Cambridge).
212
Park J. S. Chu J. S. Tsou A. D. Diop R. Tang Z. Wang A. et al . (2011). The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials32, 3921–3930. 10.1016/j.biomaterials.2011.02.019
213
Passerieux E. Rossignol R. Chopard A. Carnino A. Marini J. F. Letellier T. et al . (2006). Structural organization of the perimysium in bovine skeletal muscle: junctional plates and associated intracellular subdomains. J. Struct. Biol.154, 206–216. 10.1016/j.jsb.2006.01.002
214
Patrick C. W. Jr Chauvin P. B. Hobley J. Reece G. P. (1999). Preadipocyte seeded PLGA scaffolds for adipose tissue engineering. Tissue Eng.5, 139–151. 10.1089/ten.1999.5.139
215
Pawlikowski B. Vogler T. O. Gadek K. Olwin B. B. (2017). Regulation of skeletal muscle stem cells by fibroblast growth factors. Dev. Dyn.246, 359–367. 10.1002/dvdy.24495
216
Pearson A. M. Dutson T. R. (eds.) (1994). Quality Attributes and their Measurement in Meat, Poultry and Fish Products. Boston, MA: Springer.
217
Péault B. Rudnicki M. Torrente Y. Cossu G. Tremblay J. P. Partridge T. et al . (2007). Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol. Ther.15, 867–877. 10.1038/mt.sj.6300145
218
Pena R. N. Quintanilla R. Manunza A. Gallardo D. Casellas J. Amills M. (2014). Application of the microarray technology to the transcriptional analysis of muscle phenotypes in pigs. Anim. Genet.45, 311–321. 10.1111/age.12146
219
Perry L. Ben-Shaul S. Landau S. Levenberg S. (2017a). Co-Culture Systems for Vasculogenesis, in Vascularization for Tissue Engineering and Regenerative Medicine, eds. HolnthonerW.BanfiA.KirkpatrickJ.RedlH. (Cham: Springer International Publishing), 1–29. 10.1007/978-3-319-21056-8_7-1
220
Perry L. Flugelman M. Y. Levenberg S. (2017b). Elderly patient-derived endothelial cells for vascularization of engineered muscle. Mol. Ther.25, 935–948. 10.1016/j.ymthe.2017.02.011
221
Peters C. J. Wilkins J. L. Fick G. W. (2007). Testing a complete-diet model for estimating the land resource requirements of food consumption and agricultural carrying capacity: The New York State example. Renew. Agric. Food Syst.22, 145–153. 10.1017/S1742170507001767
222
Picard B. Berri C. Lefaucheur L. Molette C. Sayd T. Terlouw C. (2010). Skeletal muscle proteomics in livestock production. Brief. Funct. Genomics9, 259–278. 10.1093/bfgp/elq005
223
Picard B. Cassar-Malek I. Guillemin N. Bonnet M. (2011). 4.32 - Quest for Novel Muscle Pathway Biomarkers by Proteomics in Beef Production, in Comprehensive Biotechnology (Second Edition), ed. M. Moo-Young (Burlington: Academic Press), 395–405. 10.1016/B978-0-08-088504-9.00297-X
224
Picard B. Lebret B. Cassar-Malek I. Liaubet L. Berri C. Le Bihan-Duval E. et al . (2015). Recent advances in omic technologies for meat quality management. Meat Sci.109, 18–26. 10.1016/j.meatsci.2015.05.003
225
Poleti M. D. Regitano L. C. A. Souza G. H. M. F. Cesar A. S. M. Simas R. C. Silva-Vignato B. et al . (2018). Longissimus dorsi muscle label-free quantitative proteomic reveals biological mechanisms associated with intramuscular fat deposition. J. Proteomics179, 30–41. 10.1016/j.jprot.2018.02.028
226
Post M. van der Weele C. (2014). Chapter 78 - Principles of tissue engineering for food, in Principles of Tissue Engineering, Fourth Edition, eds LanzaR.LangerR.VacantiJ. (Boston, MA: Academic Press), 1647–1662. 10.1016/B978-0-12-398358-9.00078-1
227
Post M. J. (2012). Cultured meat from stem cells: challenges and prospects. Meat Sci.92, 297–301. 10.1016/j.meatsci.2012.04.008
228
Post M. J. (2014). Cultured beef: medical technology to produce food. J. Sci. Food Agric.94, 1039–1041. 10.1002/jsfa.6474
229
Post M. J. Hocquette J.-F. (2017). Chapter 16 - new sources of animal proteins: cultured Meat, in New Aspects of Meat Quality, ed. P. P. Purslow (Woodhead Publishing), 425–441. 10.1016/B978-0-08-100593-4.00017-5
230
Prieto N. Roehe R. Lavín P. Batten G. Andrés S. (2009). Application of near infrared reflectance spectroscopy to predict meat and meat products quality: a review. Meat Sci.83, 175–186. 10.1016/j.meatsci.2009.04.016
231
Purslow P. P. (2005). Intramuscular connective tissue and its role in meat quality. Meat Sci.70, 435–447. 10.1016/j.meatsci.2004.06.028
232
Purslow P. P. (2010). Muscle fascia and force transmission. J. Bodyw. Mov. Ther.14, 411–417. 10.1016/j.jbmt.2010.01.005
233
Purslow P. P. (2014). New developments on the role of intramuscular connective tissue in meat toughness. Annu. Rev. Food Sci. Technol.5, 133–153. 10.1146/annurev-food-030212-182628
234
Qazi T. H. Mooney D. J. Pumberger M. Geissler S. Duda G. N. (2015). Biomaterials based strategies for skeletal muscle tissue engineering: existing technologies and future trends. Biomaterials53, 502–521. 10.1016/j.biomaterials.2015.02.110
235
Qiu F. Xie L. Ma J.-E. Luo W. Zhang L. Chao Z. et al . (2017). Lower Expression of SLC27A1 enhances intramuscular fat deposition in chicken via down-regulated fatty acid oxidation mediated by CPT1A. Front. Physiol.8:449. 10.3389/fphys.2017.00449
236
Rafii S. Butler J. M. Ding B.-S. (2016). Angiocrine functions of organ-specific endothelial cells. Nature529, 316–325. 10.1038/nature17040
237
Ramírez-Espinosa J. J. González-Dávalos L. Shimada A. Piña E. Varela-Echavarria A. Mora O. (2016). Bovine (Bos taurus) bone marrow mesenchymal cell differentiation to adipogenic and myogenic lineages. Cells Tissues Organs201, 51–64. 10.1159/000440878
238
Reardon S. (2017). Resistance to last-ditch antibiotic has spread farther than anticipated. Nature News. Available online at: http://www.nature.com/news/resistance-to-last-ditch-antibiotic-has-spread-farther-than-anticipated-1.22140? (accessed July 17, 2018).
239
Redaelli F. Sorbona M. Rossi F. (2017). 10 - Synthesis and processing of hydrogels for medical applications, in Bioresorbable Polymers for Biomedical Applications, eds. PeraleG.HilbornJ. (Woodhead Publishing), 205–228. 10.1016/B978-0-08-100262-9.00010-0
240
Reddy N. Yang Y. (2011). Potential of plant proteins for medical applications. Trends Biotechnol.29, 490–498. 10.1016/j.tibtech.2011.05.003
241
Reilly G. C. Engler A. J. (2010). Intrinsic extracellular matrix properties regulate stem cell differentiation. J. Biomech.43, 55–62. 10.1016/j.jbiomech.2009.09.009
242
Reverter A. Hudson N. J. Wang Y. Tan S.-H. Barris W. Byrne K. A. et al . (2006). A gene coexpression network for bovine skeletal muscle inferred from microarray data. Physiol. Genomics28, 76–83. 10.1152/physiolgenomics.00105.2006
243
Roberts R. M. Yuan Y. Genovese N. Ezashi T. (2015). Livestock models for exploiting the promise of pluripotent stem cells. ILAR J.56, 74–82. 10.1093/ilar/ilv005
244
Rodriguez B. L. Larkin L. M. (2018). 12 - Functional three-dimensional scaffolds for skeletal muscle tissue engineering, in Functional 3D Tissue Engineering Scaffolds, eds. DengY.KuiperJ. (Woodhead Publishing), 279–304. 10.1016/B978-0-08-100979-6.00012-4
245
Romao J. M. Jin W. Dodson M. V. Hausman G. J. Moore S. S. Guan L. L. (2011). MicroRNA regulation in mammalian adipogenesis. Exp. Biol. Med.236, 997–1004. 10.1258/ebm.2011.011101
246
Rouwkema J. Khademhosseini A. (2016). Vascularization and angiogenesis in tissue engineering: beyond creating static networks. Trends Biotechnol.34, 733–745. 10.1016/j.tibtech.2016.03.002
247
Rouwkema J. Rivron N. C. van Blitterswijk C. A. (2008). Vascularization in tissue engineering. Trends Biotechnol.26, 434–441. 10.1016/j.tibtech.2008.04.009
248
Rowlands A. S. George P. A. Cooper-White J. J. (2008). Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am. J. Physiol. Cell Physiol.295, C1037–C1044. 10.1152/ajpcell.67.2008
249
Rowley J. A. Mooney D. J. (2002). Alginate type and RGD density control myoblast phenotype. J. Biomed. Mater. Res.60, 217–223. 10.1002/jbm.1287.abs
250
Ruel-Gariépy E. Leroux J.-C. (2004). In situ-forming hydrogels–review of temperature-sensitive systems. Eur. J. Pharm. Biopharm.58, 409–426. 10.1016/j.ejpb.2004.03.019
251
Sack M. Hofbauer A. Fischer R. Stoger E. (2015). The increasing value of plant-made proteins. Curr. Opin. Biotechnol.32, 163–170. 10.1016/j.copbio.2014.12.008
252
Sadkowski T. Ciecierska A. Majewska A. Oprzadek J. Dasiewicz K. Ollik M. et al . (2014). Transcriptional background of beef marbling—novel genes implicated in intramuscular fat deposition. Meat Sci.97, 32–41. 10.1016/j.meatsci.2013.12.017
253
Sadkowski T. Ciecierska A. Oprzadek J. Balcerek E. (2018). Breed-dependent microRNA expression in the primary culture of skeletal muscle cells subjected to myogenic differentiation. BMC Genomics19:109. 10.1186/s12864-018-4492-5
254
Sarkar N. S. Choudhury S. (2017). Algae as source of natural flavour enhancers - a mini review. Plant Sci. Today4, 172–176. 10.14719/pst.2017.4.4.338
255
Sarlio S. (2018). Towards Healthy and Sustainable Diets: Perspectives and Policy to Promote the Health of People and the Planet. Springer. Available online at: https://books.google.co.in/books?id=upRNDwAAQBAJ&redir_esc=y (accessed September 27, 2018).
256
Scarborough P. Appleby P. N. Mizdrak A. Briggs A. D. M. (2014). Dietary greenhouse gas emissions of meat-eaters, fish-eaters, vegetarians and vegans in the UK. Climate Change. Available online at: https://link.springer.com/article/10.1007/s10584-014-1169-1%EF%BB%BF (accessed September 13, 2018).
257
Schmidinger K. (2012). Worldwide Alternatives to Animal Derived Foods-Overview and Evaluation Models: Solution to Global Problems Caused by Livestock.
258
Schneider K. (2013). Concentrating on healthy feeding operations: the national school lunch program, cultured meat, and the path to a sustainable food future. J. Land Use Envtl. L.29:145.
259
Schröder M. J. A. McEachern M. G. (2004). Consumer value conflicts surrounding ethical food purchase decisions: a focus on animal welfare. Int. J. Consum. Stud.28, 168–177. 10.1111/j.1470-6431.2003.00357.x
260
Shahidi F. (2002). Lipid-derived flavors in meat products, in Meat Processing: Improving Quality, eds KerryJ.KerryJ.LedwardD. (Cambridge: Woodhead Publishing Ltd), 105–121. Available online at: https://books.google.co.in/books?id=_A-kAgAAQBAJ&redir_esc=y (accessed September 03, 2018).
261
Shahidi F. (2012). Flavor of Meat and Meat Products. Springer Science & Business Media. Available online at: https://www.springer.com/gp/book/9781461359111 (accessed September 02, 2018).
262
Shahidi F. Rubin L. J. D'Souza L. A. (1986). Meat flavor volatiles: a review of the composition, techniques of analysis, and sensory evaluation. Crit. Rev. Food Sci. Nutr.24, 141–243. 10.1080/10408398609527435
263
Shandalov Y. Egozi D. Freiman A. Rosenfeld D. Levenberg S. (2015). A method for constructing vascularized muscle flap. Methods84, 70–75. 10.1016/j.ymeth.2015.03.021
264
Shandalov Y. Egozi D. Koffler J. Dado-Rosenfeld D. Ben-Shimol D. Freiman A. et al . (2014). An engineered muscle flap for reconstruction of large soft tissue defects. Proc. Natl. Acad. Sci. U.S.A.111, 6010–6015. 10.1073/pnas.1402679111
265
Sharma S. Thind S. S. Kaur A. (2015). In vitro meat production system: why and how?J. Food Sci. Technol.52, 7599–7607. 10.1007/s13197-015-1972-3
266
Shit S. C. Shah P. M. (2014). Edible polymers: challenges and opportunities. J. Polym. Environ.2014:427259. 10.1155/2014/427259
267
Shoseyov O. Posen Y. Grynspan F. (2013). Human recombinant type I collagen produced in plants. Tissue Eng. Part A19, 1527–1533. 10.1089/ten.tea.2012.0347
268
Shoseyov O. Stein H. (2017). Collagen producing plants and methods of generating and using same. US Patent. Available online at: https://patents.google.com/patent/US9783816B2/en (accessed October 3, 2018).
269
Sims D. E. (1986). The pericyte—A review. Tissue and Cell18, 153–174. 10.1016/0040-8166(86)90026-1
270
Slade P. (2018). If you build it, will they eat it? Consumer preferences for plant-based and cultured meat burgers. Appetite125, 428–437. 10.1016/j.appet.2018.02.030
271
Smith S. B. Kawachi H. Choi C. B. Choi C. W. Wu G. Sawyer J. E. (2009). Cellular regulation of bovine intramuscular adipose tissue development and composition. J. Anim. Sci.87, E72–82. 10.2527/jas.2008-1340
272
Snell K. D. Singh V. Brumbley S. M. (2015). Production of novel biopolymers in plants: recent technological advances and future prospects. Curr. Opin. Biotechnol.32, 68–75. 10.1016/j.copbio.2014.11.005
273
Specht E. A. Welch D. R. Rees Clayton E. M. Lagally C. D. (2018). Opportunities for applying biomedical production and manufacturing methods to the development of the clean meat industry. Biochem. Eng. J.132, 161–168. 10.1016/j.bej.2018.01.015
274
Specht L. (2019). An Analysis of Culture Medium Costs and Production Volumes for Cell-Based Meat. Available online at: https://www.gfi.org/files/sci-tech/clean-meat-production-volume-and-medium-cost.pdf
275
Steinfeld H. Gerber P. Wassenaar T. Castel V. Rosales M. Haan C. et al . (2006). Livestock's Long Shadow: Environmental Issues and Options. Food and Agriculture Organization of the United Nations (FAO) Available online at: http://www.fao.org/3/a0701e/a0701e00.htm (accessed August 31, 2015).
276
Stephens N. Di Silvio L. Dunsford I. Ellis M. Glencross A. Sexton A. (2018a). Bringing cultured meat to market: Technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci. Technol.78, 155–166. 10.1016/j.tifs.2018.04.010
277
Stephens N. King E. Lyall C. (2018b). Blood, meat, and upscaling tissue engineering: Promises, anticipated markets, and performativity in the biomedical and agri-food sectors. Biosocieties13, 368–388. 10.1057/s41292-017-0072-1
278
Sun X. Li M. Sun Y. Cai H. Lan X. Huang Y. et al . (2016). The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. Biochim. Biophys. Acta1863, 2835–2845. 10.1016/j.bbamcr.2016.08.014
279
Sweeney S. M. Orgel J. P. Fertala A. McAuliffe J. D. Turner K. R. Di Lullo G. A. et al . (2008). Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J. Biol. Chem.283, 21187–21197. 10.1074/jbc.M709319200
280
Syverud B. C. Lee J. D. VanDusen K. W. Larkin L. M. (2014). Isolation and Purification of satellite cells for skeletal muscle tissue engineering. J. Regen. Med. Tissue Eng.3:117. 10.4172/2325-9620.1000117
281
Sze J. H. Brownlie J. C. Love C. A. (2016). Biotechnological production of hyaluronic acid: a mini review. Biotech6:67. 10.1007/s13205-016-0379-9
282
Takenouchi T. Miyashita N. Ozutsumi K. Rose M. T. Aso H. (2004). Role of caveolin-1 and cytoskeletal proteins, actin and vimentin, in adipogenesis of bovine intramuscular preadipocyte cells. Cell Biol. Int.28, 615–623. 10.1016/j.cellbi.2004.05.003
283
Tan S. H. Reverter A. Wang Y. Byrne K. A. McWilliam S. M. Lehnert S. A. (2006). Gene expression profiling of bovine in vitro adipogenesis using a cDNA microarray. Funct. Integr. Genomics6, 235–249. 10.1007/s10142-005-0016-x
284
Tang K. L. Caffrey N. P. Nóbrega D. B. Cork S. C. Ronksley P. E. Barkema H. W. et al . (2017). Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health1, e316–e327. 10.1016/S2542-5196(17)30141-9
285
Taniguchi M. Guan L. L. Zhang B. Dodson M. V. Okine E. Moore S. S. (2008). Adipogenesis of bovine perimuscular preadipocytes. Biochem. Biophys. Res. Commun.366, 54–59. 10.1016/j.bbrc.2007.11.110
286
Tedesco F. S. Moyle L. A. Perdiguero E. (2017). Muscle Interstitial Cells: A Brief Field Guide to Non-satellite Cell Populations in Skeletal Muscle, in Muscle Stem Cells: Methods and Protocols, eds. PerdigueroE.CornelisonD. D. W. (New York, NY: Springer), 129–147. 10.1007/978-1-4939-6771-1_7
287
Thornton K. J. Kamange-Sollo E. White M. E. Dayton W. R. (2015). Role of G protein-coupled receptors (GPCR), matrix metalloproteinases 2 and 9 (MMP2 and MMP9), heparin-binding epidermal growth factor-like growth factor (hbEGF), epidermal growth factor receptor (EGFR), erbB2, and insulin-like growth factor 1 receptor (IGF-1R) in trenbolone acetate-stimulated bovine satellite cell proliferation. J. Anim. Sci.93, 4291–4301. 10.2527/jas.2015-9191
288
Thorsteinsdóttir S. Deries M. Cachaço A. S. Bajanca F. (2011). The extracellular matrix dimension of skeletal muscle development. Dev. Biol.354, 191–207. 10.1016/j.ydbio.2011.03.015
289
Tizioto P. C. Coutinho L. L. Mourão G. B. Gasparin G. Malagó W. Jr Bressani F. A. et al . (2016). Variation in myogenic differentiation 1 mRNA abundance is associated with beef tenderness in Nelore cattle. Anim. Genet.47, 491–494. 10.1111/age.12434
290
Tong J. Zhu M. J. Underwood K. R. Hess B. W. Ford S. P. Du M. (2008). AMP-activated protein kinase and adipogenesis in sheep fetal skeletal muscle and 3T3-L1 cells. J. Anim. Sci.86, 1296–1305. 10.2527/jas.2007-0794
291
Trivedi D. K. Hollywood K. A. Rattray N. J. W. Ward H. Trivedi D. K. Greenwood J. et al . (2016). Meat, the metabolites: an integrated metabolite profiling and lipidomics approach for the detection of the adulteration of beef with pork. Analyst141, 2155–2164. 10.1039/C6AN00108D
292
Tse J. R. Engler A. J. (2011). Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS ONE6:e15978. 10.1371/journal.pone.0015978
293
Tuomisto H. L. de Mattos M. J. T. (2011). Environmental impacts of cultured meat production. Environ. Sci. Technol.45, 6117–6123. 10.1021/es200130u
294
United States Department of Agriculture (2018). Livestock, Poultry, & Grain Market News. Available online at: https://www.ams.usda.gov/mnreports/pywretailchicken.pdf (accessed September 12, 2018).
295
Van der Gucht O. (2018). Cultured Meat: Current State of the Art and Future Challenges. Available online at: https://lib.ugent.be/fulltxt/RUG01/002/481/330/RUG01-002481330_2018_0001_AC.pdf
296
van der Weele C. Driessen C. (2013). Emerging profiles for cultured meat; ethics through and as design. Animals3, 647–662. 10.3390/ani3030647
297
van Hinsbergh V. W. Collen A. Koolwijk P. (2001). Role of fibrin matrix in angiogenesis. Ann. N. Y. Acad. Sci.936, 426–437. 10.1111/j.1749-6632.2001.tb03526.x
298
Varzaneh F. E. Shillabeer G. Wong K. L. Lau D. C. (1994). Extracellular matrix components secreted by microvascular endothelial cells stimulate preadipocyte differentiation in vitro. Metabolism43, 906–912. 10.1016/0026-0495(94)90275-5
299
Veiseth-Kent E. de Almeida A. M. Picard B. Hollung K. (2018). Proteomics in skeletal muscle research, in Proteomics in Domestic Animals: From Farm to Systems Biology, eds. de AlmeidaA. M.EckersallD.MillerI. (Cham: Springer International Publishing), 195–217. 10.1007/978-3-319-69682-9_10
300
Veliça P. Bunce C. M. (2011). A quick, simple and unbiased method to quantify C2C12 myogenic differentiation. Muscle Nerve44, 366–370. 10.1002/mus.22056
301
Verbeke W. Marcu A. Rutsaert P. Gaspar R. Seibt B. Fletcher D. et al . (2015). Would you eat cultured meat?” Consumers' reactions and attitude formation in Belgium, Portugal and the United Kingdom. Meat Sci.102, 49–58. 10.1016/j.meatsci.2014.11.013
302
Verbeke W. A. J. Viaene J. (2000). Ethical challenges for livestock production:meeting consumer concerns about meat safety and animalwelfare. J. Agric. Environ. Ethics12, 141–151. 10.1023/A:1009538613588
303
Verbruggen S. Luining D. van Essen A. Post M. J. (2018). Bovine myoblast cell production in a microcarriers-based system. Cytotechnology70, 503–512. 10.1007/s10616-017-0101-8
304
Vitello L. Radu C. Malerba A. Segat D. Cantini M. Carraro U. et al . (2004). Enhancing myoblast proliferation by using myogenic factors: a promising approach for improving fiber regeneration in sport medicine and skeletal muscle diseases. Basic Appl. Myol.14, 45–51.
305
Volz A.-C. Huber B. Kluger P. J. (2016). Adipose-derived stem cell differentiation as a basic tool for vascularized adipose tissue engineering. Differentiation92, 52–64. 10.1016/j.diff.2016.02.003
306
Von Heimburg D. Serov G. Oepen T. Pallua N. (2003). Fat tissue engineering. Topics in Tissue Engineering. Available online at: http://www.oulu.fi/spareparts/ebook_topics_in_t_e/abstracts/heimburg_01.pdf.
307
von Tell D. Armulik A. Betsholtz C. (2006). Pericytes and vascular stability. Exp. Cell Res.312, 623–629. 10.1016/j.yexcr.2005.10.019
308
Wagner W. Horn P. Castoldi M. Diehlmann A. Bork S. Saffrich R. et al . (2008). Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS ONE3:e2213. 10.1371/journal.pone.0002213
309
Wang G. Kim W. K. Cline M. A. Gilbert E. R. (2017a). Factors affecting adipose tissue development in chickens: a review. Poult. Sci.96, 3687–3699. 10.3382/ps/pex184
310
Wang T. Lew J. Premkumar J. Poh C. L. Naing M. W. (2017b). Production of recombinant collagen: state of the art and challenges. Eng. Biol.1, 18–23. 10.1049/enb.2017.0003
311
Wang Y.-H. Byrne K. A. Reverter A. Harper G. S. Taniguchi M. McWilliam S. M. et al . (2005). Transcriptional profiling of skeletal muscle tissue from two breeds of cattle. Mamm. Genome16, 201–210. 10.1007/s00335-004-2419-8
312
Wang Y. H. Bower N. I. Reverter A. Tan S. H. De Jager N. Wang R. et al . (2009). Gene expression patterns during intramuscular fat development in cattle1. J. Ani. Sci.87, 119–130. 10.2527/jas.2008-1082
313
Wang Y. M. Ding X. B. Dai Y. Liu X. F. Guo H. Zhang Y. (2015). Identification and bioinformatics analysis of miRNAs involved in bovine skeletal muscle satellite cell myogenic differentiation. Mol. Cell. Biochem.404, 113–122. 10.1007/s11010-015-2371-9
314
Warriss P. D. Rhodes D. N. (1977). Haemoglobin concentrations in beef. J. Sci. Food Agric.28, 931–934. 10.1002/jsfa.2740281012
315
Webster A. J. (2001). Farm animal welfare: the five freedoms and the free market. Vet. J.161, 229–237. 10.1053/tvjl.2000.0563
316
Webster J. (2006). Animal sentience and animal welfare: What is it to them and what is it to us?Appl. Anim. Behav. Sci.100, 1–3. 10.1016/j.applanim.2006.05.012
317
Werkmeister J. A. Ramshaw J. A. M. (2012). Recombinant protein scaffolds for tissue engineering. Biomed. Mater.7:012002. 10.1088/1748-6041/7/1/012002
318
Wickramasinghe S. Cánovas A. Rincón G. Medrano J. F. (2014). RNA-Sequencing: A tool to explore new frontiers in animal genetics. Livest. Sci.166, 206–216. 10.1016/j.livsci.2014.06.015
319
Wilfred B. R. Wang W.-X. Nelson P. T. (2007). Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab.91, 209–217. 10.1016/j.ymgme.2007.03.011
320
Will K. Schering L. Albrecht E. Kalbe C. Maak S. (2015). Differentiation of bovine satellite cell-derived myoblasts under different culture conditions. In Vitro Cell. Dev. Biol. Anim.51, 885–889. 10.1007/s11626-015-9916-9
321
Wittmann K. Dietl S. Ludwig N. Berberich O. Hoefner C. Storck K. et al . (2015). Engineering vascularized adipose tissue using the stromal-vascular fraction and fibrin hydrogels. Tissue Eng. Part A21, 1343–1353. 10.1089/ten.tea.2014.0299
322
Woerdeman D. L. Ye P. Shenoy S. Parnas R. S. Wnek G. E. Trofimova O. (2005). Electrospun fibers from wheat protein: investigation of the interplay between molecular structure and the fluid dynamics of the electrospinning process. Biomacromolecules6, 707–712. 10.1021/bm0494545
323
Wolf M. T. Dearth C. L. Sonnenberg S. B. Loboa E. G. Badylak S. F. (2015). Naturally derived and synthetic scaffolds for skeletal muscle reconstruction. Adv. Drug Deliv. Rev.84, 208–221. 10.1016/j.addr.2014.08.011
324
Won S. Jung J. Park E. Kim H. (2018). Identification of genes related to intramuscular fat content of pigs using genome-wide association study. Asian-australas. J. Anim. Sci.31, 157–162. 10.5713/ajas.17.0218
325
Wood J. D. Enser M. Fisher A. V. Nute G. R. Sheard P. R. Richardson R. I. et al . (2008). Fat deposition, fatty acid composition and meat quality: a review. Meat Sci.78, 343–358. 10.1016/j.meatsci.2007.07.019
326
Wood J. D. Richardson R. I. Nute G. R. Fisher A. V. Campo M. M. Kasapidou E. et al . (2004). Effects of fatty acids on meat quality: a review. Meat Sci.66, 21–32. 10.1016/S0309-1740(03)00022-6
327
Woodard S. L. Mayor J. M. Bailey M. R. Barker D. K. Love R. T. Lane J. R. et al . (2003). Maize (Zea mays)-derived bovine trypsin: characterization of the first large-scale, commercial protein product from transgenic plants. Biotechnol. Appl. Biochem.38, 123–130. 10.1042/BA20030026
328
Xu J. Towler M. Weathers P. J. (2016). Platforms for plant-based protein production, in Bioprocessing of Plant In Vitro Systems, eds. PavlovA.BleyT. (Cham: Springer International Publishing), 1–40. 10.1007/978-3-319-32004-5_14-1
329
Yan X. Zhu M.-J. Dodson M. V. Du M. (2013). Developmental programming of fetal skeletal muscle and adipose tissue development. J. Genomics1, 29–38. 10.7150/jgen.3930
330
Yang S. Xin X. Zlot C. Ingle G. Fuh G. Li B. et al . (2001). Vascular endothelial cell growth factor-driven endothelial tube formation is mediated by vascular endothelial cell growth factor receptor-2, a kinase insert domain-containing receptor. Arterioscler. Thromb. Vasc. Biol.21, 1934–1940. 10.1161/hq1201.099432
331
Yegappan R. Selvaprithiviraj V. Amirthalingam S. Jayakumar R. (2018). Carrageenan based hydrogels for drug delivery, tissue engineering and wound healing. Carbohydr. Polym.198, 385–400. 10.1016/j.carbpol.2018.06.086
332
Yin H. Price F. Rudnicki M. A. (2013). Satellite cells and the muscle stem cell niche. Physiol. Rev.93, 23–67. 10.1152/physrev.00043.2011
333
Young D. A. Choi Y. S. Engler A. J. Christman K. L. (2013). Stimulation of adipogenesis of adult adipose-derived stem cells using substrates that mimic the stiffness of adipose tissue. Biomaterials34, 8581–8588. 10.1016/j.biomaterials.2013.07.103
334
Yuan Y. (2018). Capturing bovine pluripotency. Proc. Natl. Acad. Sci. U. S. A.115, 1962–1963. 10.1073/pnas.1800248115
335
Zahoor I. de Koning D.-J. Hocking P. M. (2017). Transcriptional profile of breast muscle in heat stressed layers is similar to that of broiler chickens at control temperature. Genet. Sel. Evol.49:69. 10.1186/s12711-017-0346-x
336
Zammit P. S. Partridge T. A. Yablonka-Reuveni Z. (2006). The skeletal muscle satellite cell: the stem cell that came in from the cold. J. Histochem. Cytochem.54, 1177–1191. 10.1369/jhc.6R6995.2006
337
Zeltinger J. Sherwood J. K. Graham D. A. Müeller R. Griffith L. G. (2001). Effect of pore size and void fraction on cellular adhesion, proliferation, and matrix deposition. Tissue Eng.7, 557–572. 10.1089/107632701753213183
338
Zhang G. H. Lu J. X. Chen Y. Zhao Y. Q. Guo P. H. Yang J. T. et al . (2014). Comparison of the adipogenesis in intramuscular and subcutaneous adipocytes from Bamei and Landrace pigs. Biochem. Cell Biol.92, 259–267. 10.1139/bcb-2014-0019
339
Zhang W. R. Zhang H. N. Wang Y. M. Dai Y. Liu X. F. Li X. et al . (2017). miR-143 regulates proliferation and differentiation of bovine skeletal muscle satellite cells by targeting IGFBP5. In Vitro Cell. Dev. Biol. Anim.53, 265–271. 10.1007/s11626-016-0109-y
340
Zhou G. Wang X. Yuan C. Kang D. Xu X. Zhou J. et al . (2017). Integrating miRNA and mRNA expression profiling uncovers miRNAs underlying fat deposition in sheep. Biomed Res. Int.2017:1857580. 10.1155/2017/1857580
Summary
Keywords
tissue engineering, skeletal muscle, clean meat, cell-based meat, cultured meat, cellular agriculture, cell culture
Citation
Ben-Arye T and Levenberg S (2019) Tissue Engineering for Clean Meat Production. Front. Sustain. Food Syst. 3:46. doi: 10.3389/fsufs.2019.00046
Received
06 November 2018
Accepted
29 May 2019
Published
18 June 2019
Volume
3 - 2019
Edited by
Mark J. Post, Maastricht University, Netherlands
Reviewed by
Sara M. Oliveira, Laboratório Ibérico Internacional de Nanotecnologia (INL), Portugal; Lorenzo Moroni, Maastricht University, Netherlands
Updates
Copyright
© 2019 Ben-Arye and Levenberg.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shulamit Levenberg shulamit@bm.technion.ac.il
This article was submitted to Sustainable Food Processing, a section of the journal Frontiers in Sustainable Food Systems
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.