- 1Department of Pure Mathematics, Lecturer of Mathematics at Architecture and Computer Engineering Department, University of Applied Sciences and Technology (Unit 10), Tehran, Iran
- 2School of Cardiovascular and Metabolic Health, University of Glasgow, Glasgow, United Kingdom
- 3Human Performance Laboratory, Education, Physical Activity and Health Research Unit, Universidad Católica del Maule, Talca, Chile
- 4Colorectal Research Center, Iran University of Medical Sciences, Tehran, Iran
- 5British Heart Foundation Cardiovascular Research Centre, University of Glasgow, Glasgow, United Kingdom
- 6Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 7Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Background: The increasing prevalence of colorectal cancer (CRC) in Iran over the past three decades has made it a key public health burden. This study aimed to predict metastasis in CRC patients using machine learning (ML) approaches in terms of demographic and clinical factors.
Methods: This study focuses on 1,127 CRC patients who underwent appropriate treatments at Taleghani Hospital, a tertiary care facility. The patients were divided into training and test datasets in an 80:20 ratio. Various ML methods, including Naive Bayes (NB), random rorest (RF), support vector machine (SVM), neural network (NN), decision tree (DT), and logistic regression (LR), were used for predicting metastasis in CRC patients. Model performance was evaluated using 5-fold cross-validation, reporting sensitivity, specificity, the area under the curve (AUC), and other indexes.
Results: Among the 1,127 patients, 183 (16%) had experienced metastasis. In the predictionof metastasis, both the NN and RF algorithms had the highest AUC, while SVM ranked third in both the original and balanced datasets. The NN and RF algorithms achieved the highest AUC (100%), sensitivity (100% and 100%, respectively), and accuracy (99.2% and 99.3%, respectively) on the balanced dataset, followed by the SVM with an AUC of 98.8%, a sensitivity of 97.5%, and an accuracy of 97%. Moreover, lower false negative rate (FNR), false positive rate (FPR), and higher negative predictive value (NPV) can be confirmed by these two methods. The results also showed that all methods exhibited good performance in the test datasets, and the balanced dataset improved the performance of most ML methods. The most important variables for predicting metastasis were the tumor stage, the number of involved lymph nodes, and the treatment type. In a separate analysis of patients with tumor stages I–III, it was identified that tumor grade, tumor size, and tumor stage are the most important features.
Conclusion: This study indicated that NN and RF were the best among ML-based approaches for predicting metastasis in CRC patients. Both the tumor stage and the number of involved lymph nodes were considered the most important features.
Background
Colorectal cancer (CRC) has been regarded as one of the four most common types of cancer as well as the second leading cause of cancer deaths (Siegel et al., 2016). While there have been promising advancements in reducing the incidence of CRC, both morbidity and mortality rates remain high (Wieszczy et al., 2020). CRC has been steadily increasing worldwide since the 1960s, with mortality rates varying significantly according to geographical locations (Ferlay et al., 2019).
Machine learning (ML) models have become essential tools for identifying individuals at an increased risk of developing colorectal cancer and uncovering risk factors associated with the disease (Kourou et al., 2015). The ML algorithms have revealed magnificent performance in predicting survival cancers and their metastasis history. The ML-based approaches represent innovative and practical models for effectively predicting overall survival (OS) among CRC patients (Manilich et al., 2011; Wen et al., 2021). The ML methods overcome challenges in estimating coefficients and accurately modeling data. Additionally, ML models can automatically handle noise in datasets, non-linearity, complex interactions, large sample sizes, and numerous features. Overall, ML approaches have shown promise in improving treatment outcomes in cancer research (Zhou et al., 2021; Greener et al., 2022; Talebi et al., 2023). ML-based approaches have also been employed to predict metastatic relapse in breast cancer in several studies (Tapak et al., 2019; Nicol et al., 2020). Furthermore, numerous epidemiological studies have investigated specific hypotheses related to CRC risk factors (Talebi et al., 2019, 2020; Borumandnia et al., 2021). These studies encompass survival analysis techniques such as Cox proportional hazards, time-dependent Cox, cure models, and other types of survival analysis using clinical datasets. Moreover, other investigations have utilized ML models such as decision tree (DT), support vector machine (SVM), neural network (NN), and Naive Bayes (NB) methods (Talebi et al., 2023).
This historical cohort study aims to predict CRC survival using supervised machine learning methods, including NB, random forest (RF), SVM, NN, DT, and logistic regression (LR).
Methods
Clinical data from 1,127 patients who underwent medical treatment for rectal cancer at Taleghani Hospital, a tertiary care facility, from 2013 to 2019, were scrutinized to develop prediction models using an ML classifier. Metastasis served as the dependent variable, while demographic and clinical factors were considered as independent variables. Demographic characteristics included sex, age, education level, smoking, marital status, and BMI, while clinical factors encompassed the number of involved lymph nodes, tumor grade, tumor stage, treatment type, and diabetes mellitus.
This research adheres to the principles of the Helsinki Declaration. The methods were conducted in accordance with relevant guidelines and official instructions. The ethics committee of Iran University of Medical Sciences has waived the requirement for informed consent and approved the project (IR.IUMS.REC.1400.459).
Inclusion and exclusion criteria
The inclusion criteria for the CRC screening program, which spanned from 2013 to 2019, included various diagnostic methods such as endoscopy, imaging, stool, or blood tests, and colonoscopy reports for patients diagnosed with CRC from 2013 to 2019. Additionally, the clinical data were utilized for assessing the overall or relative survival of CRC patients who had experienced metastasis. On the contrary, the exclusion criteria included the absence of a colonoscopy report or incomplete data.
Preliminary processing of data
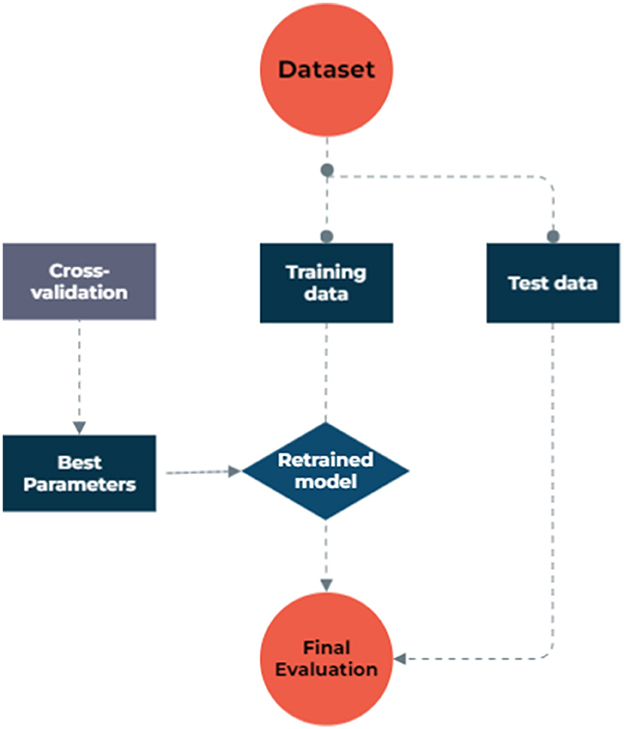
To address missing data, we utilized model-based imputation techniques. The dataset used in the study consisted of 1,127 samples and 13 factors, including patients' demographic and clinical characteristics, as well as their metastasis status as an outcome binary variable, obtained from archived records. To facilitate the analysis, categorical features were transformed into discrete values using binning discretization methods. The continuous variables were normalized through one-hot encoding, centering them around the mean and scaling to a standard deviation of 1. In the NB model process, we grouped numeric values into four equally frequent categories. The dataset exhibited a significantly imbalanced distribution of metastasis rates, with a ratio of 16 metastasis to 84 non-metastasis cases, which could lead to biased models that favor the majority class and ignore the minority class, resulting in poor sensitivity and precision. To overcome this issue, the synthetic minority oversampling technique (SMOTE) was employed to balance the data by creating synthetic samples from the minority class. The SMOTE created synthetic samples from the minority class by finding the k-nearest neighbors of each sample and randomly choosing one of them to create a new sample along the line connecting them, which led to a new balanced dataset. Both the original and balanced datasets were then used to implement various ML algorithms, and their results were compared in terms of model performances. Figure 1 illustrates the step-by-step data processing and model selection process.
Model development
ML models were developed using the five-fold cross-validation method to assess the impact of the selected variables. The dataset was divided into five randomly selected folds, with 4-folds used for model training and the remaining fold applied for testing. This process ensured that 80% of the data was used for training and 20% for testing in each iteration. Various algorithms, including NB, RF, SVM, NN, DT, and LR, were employed to create models for predicting metastasis in CRC patients. The hyperparameters were tuned using a grid search technique, which involves testing different combinations of hyperparameter values and selecting the best one based on the cross-validation performance and prior experience. The performance of the models was evaluated using the validation data, and this iterative process was repeated until satisfactory results were obtained. In the case of the NN approach, algorithm selection was done through trial and error. A multi-layer perceptron network with the rectified linear unit activation function and stochastic gradient-based optimizer for weight optimization was utilized for NN modeling. For DT, a forward pruning technique was employed to split the data based on class purity. The RF algorithm constructed a set of decision trees using bootstrap sampling from the training data. The SVM algorithm employed a radial basis function kernel in this study. The modeling process for SVM included setting the cost to 1.00, regression loss to 0.1, and numerical tolerance to 0.001. Additionally, an NB classifier based on the Bayes' theorem was fitted. This method is known for its speed and robust performance.
Statistical analysis
The characteristics of participants were presented by reporting the mean ± SD for continuous variables and frequency with percentage for categorical ones. Missing data were imputed using model-based imputation methods. The ML-based approaches, including NB, RF, SVM, NN, DT, and LR, were used for predicting metastasis in CRC patients. The data were divided into training and test datasets in an 80:20 ratio. Then, the performance of various ML methods on both the original and balanced datasets was compared using 5-fold cross-validation and ROC curves. The area under the curve (AUC) of ROC, Precision–Recall area under the curve (PR-AUC), sensitivity, specificity, false negative rate (FNR), false positive rate (FPV), negative predictive value (NPV), F1 score, accuracy, and precision were reported for both the training and test sets in both the original and balanced datasets. The training accuracy is the overall accuracy of the model obtained by averaging the accuracies from the individual cross-validation runs. Additionally, calculating the performance analysis and obtaining the vital factors were carried out in stages I, II, and III. The ML modeling was implemented using Orange3 software version 3.36.1 and R studio version 4.2.0.
Results
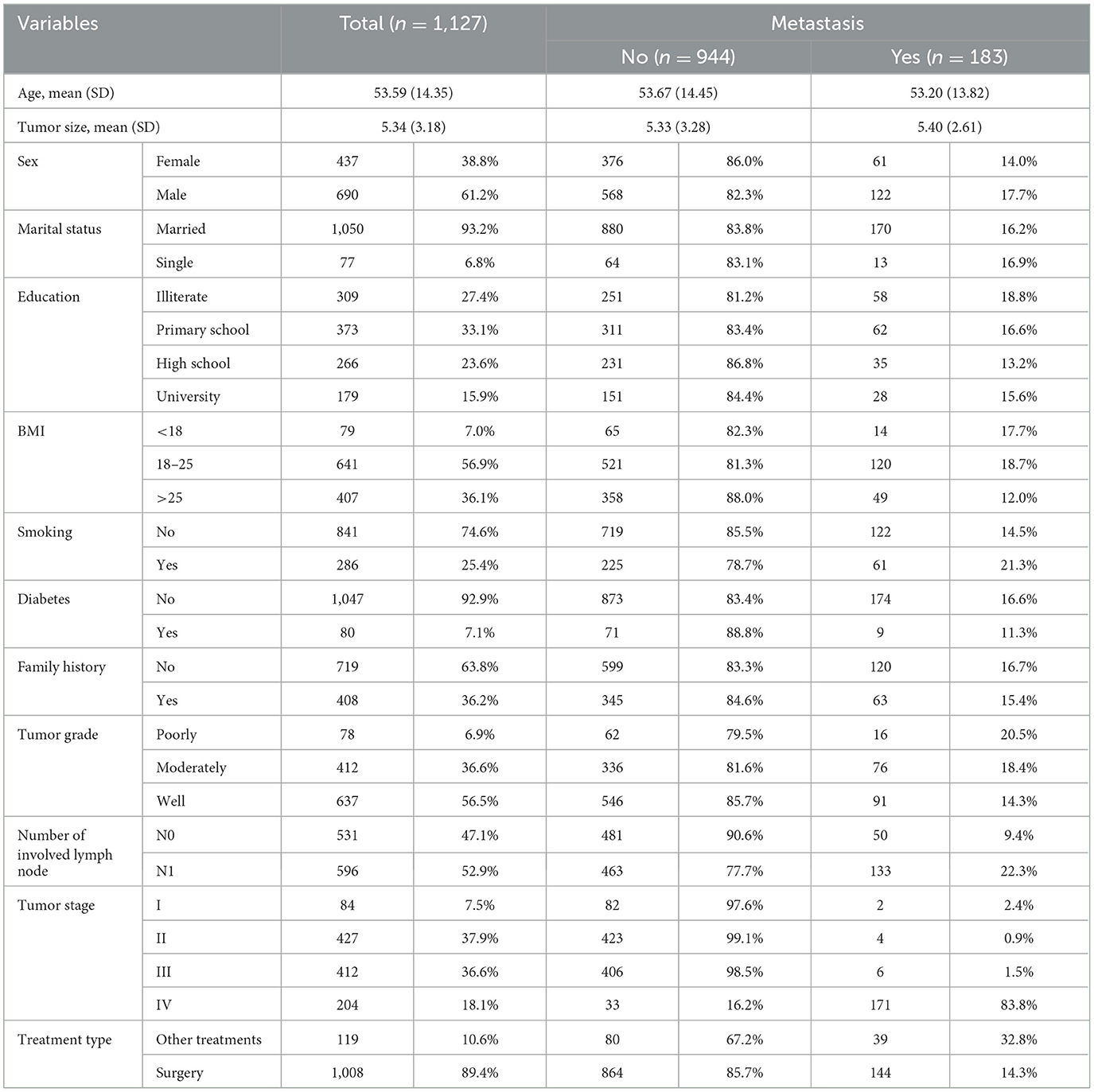
A total of 1,127 registered CRC patients who experienced metastasis in 183 (16.2%) cases were included in this historical cohort study. The mean ± SD age of patients was 53.59 ± 14.35 years, ranging from 14 to 94 years. Out of the total number of patients, 437 (38.8%) patients were women. Demographic and clinical information is provided in Table 1.
Subsequently, various ML algorithms were applied to predict metastasis in CRC patients. Table 2 reveals the performance of different ML algorithms, which were evaluated using a 5-fold CV. It can be explained that the performance of all methods was acceptable in clinical research. The highest sensitivity and specificity (97.9% and 98.7%, respectively) were estimated for the NN algorithm. Both NN and RF algorithms had the highest AUC, while the SVM ranked third in both the original and balanced datasets. In addition, the evaluation of the results indicated that all methods exhibited good performance in the test datasets. The results also showed that the balanced dataset improved the performance of most ML methods, especially DT and NN, in predicting metastasis in CRC patients. As shown in Table 2, the balanced dataset increased the sensitivity and AUC of most ML methods, indicating that the models were able to better distinguish between metastasis and non-metastasis cases. The NN and RF achieved the highest AUC (100%), sensitivity (100% and 100%, respectively), and accuracy (99.2% and 99.3%, respectively) on the balanced dataset, followed by the SVM with an AUC of 98.8%, a sensitivity of 97.5%, and an accuracy of 97%. Moreover, lower FNR and FPR and higher NPV can be confirmed by these two methods. These results suggest that the NN and RF models are the most suitable ML methods for predicting metastasis in CRC patients as they can capture the complex and non-linear relationships between the features and the outcome. The SVM, LR, and NB models also showed improved performance on the balanced dataset, while they were still inferior in the NN and RF models.
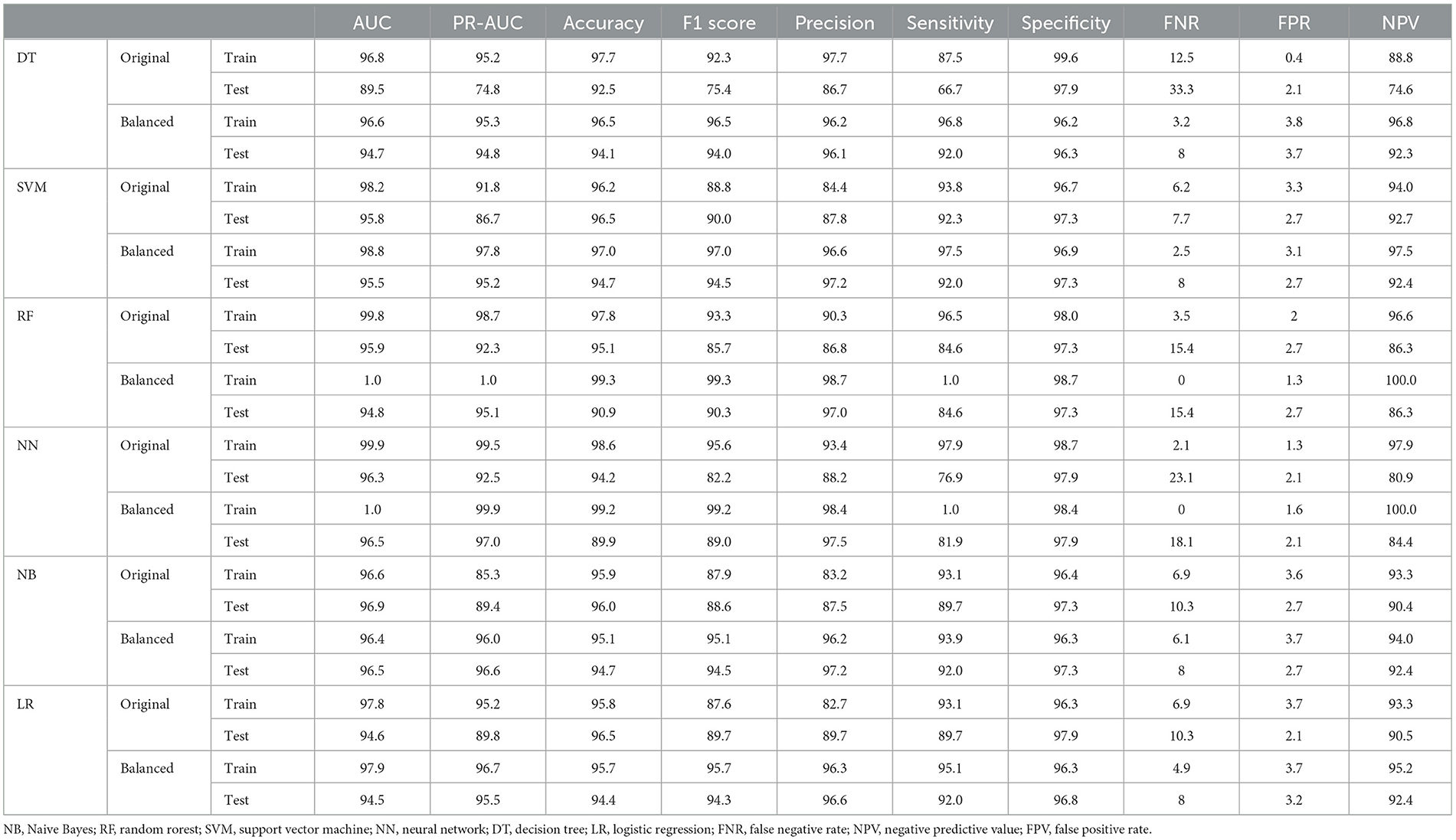
Table 2. Performance criteria for the ML methods for predicting metastasis in colorectal cancer patients.
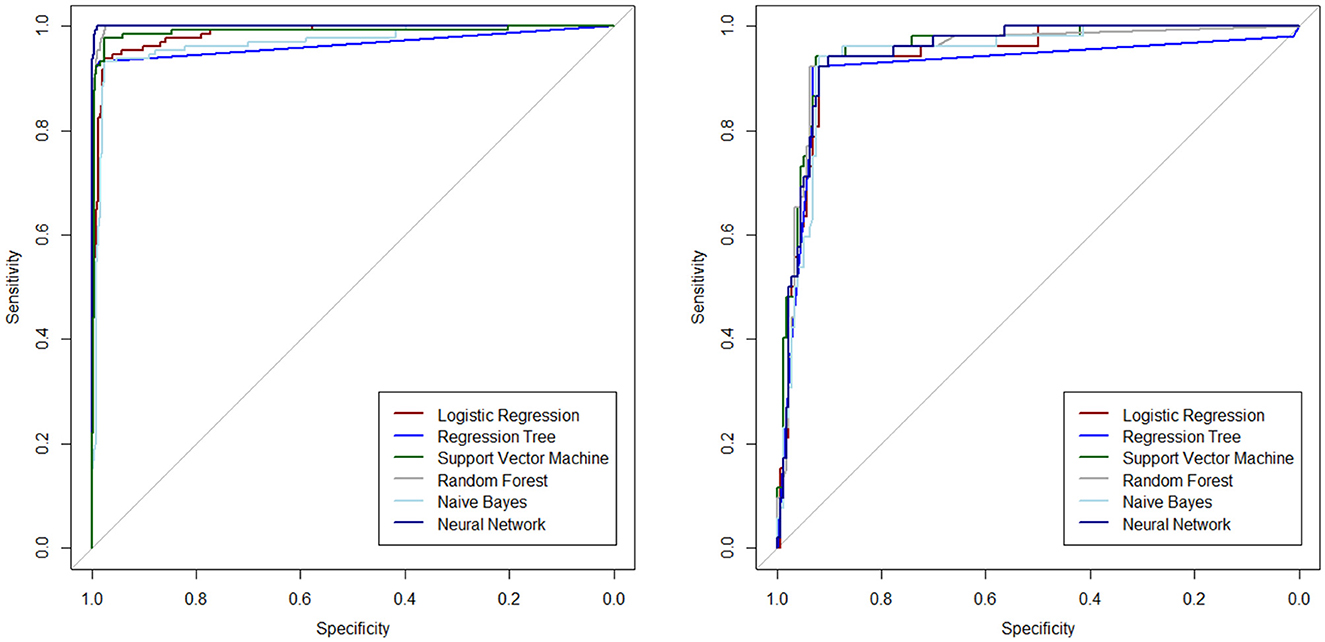
The ROC curves are plotted to determine the diagnostic ability of the ML algorithms in Figure 2, separately for the test and training datasets.
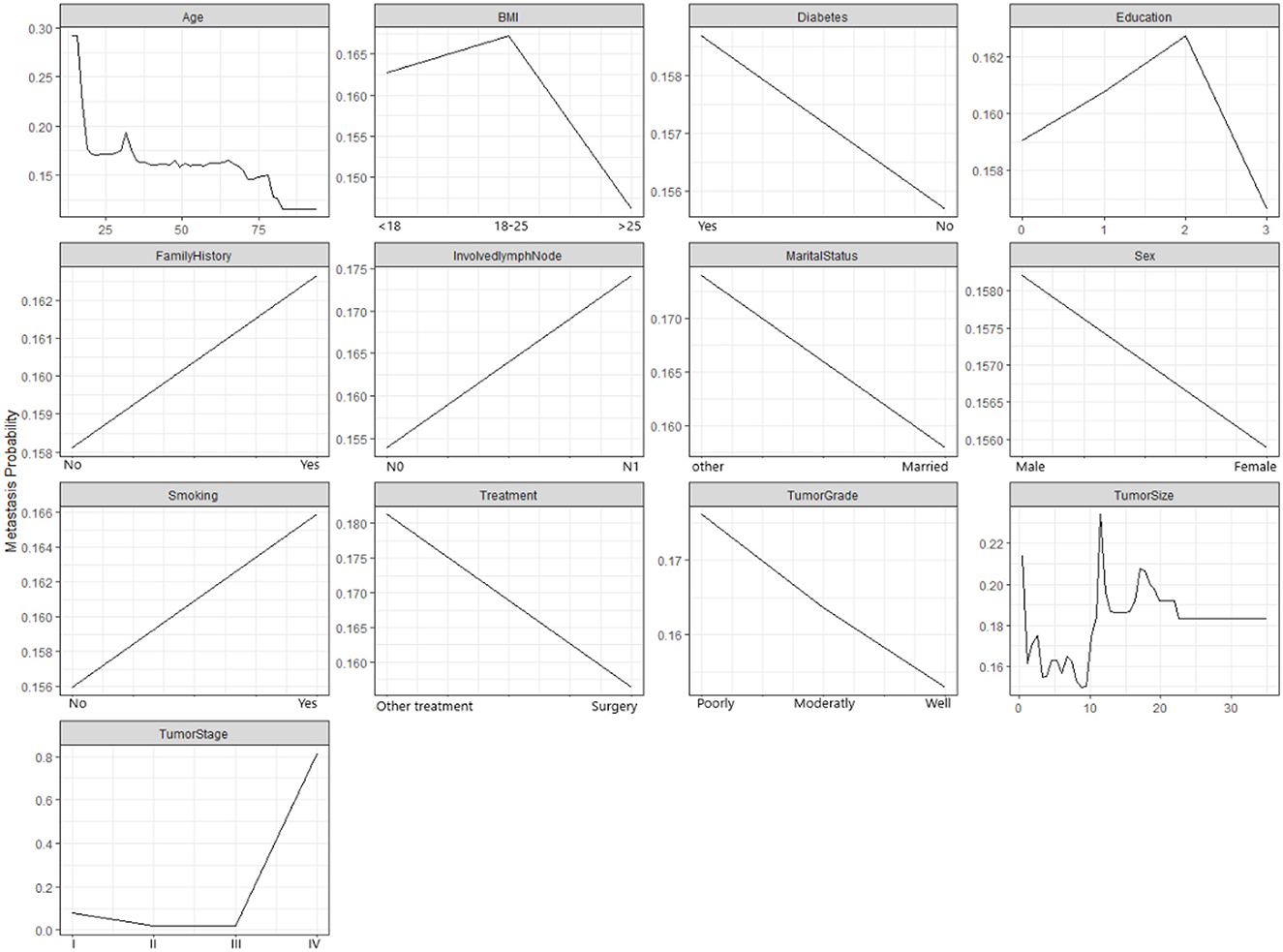
Figure 3 demonstrates how the probability of metastasis depends on various features of the patients and their tumors. Some of the graphs show a clear relationship between the feature and the probability of metastasis, such as age, tumor size, and tumor stage. For example, the graph for the tumor stage shows that the probability of metastasis increases as the the tumor stage increases, which means that more advanced tumors are more likely to spread than less advanced tumors. Some of the graphs show a weak or unclear relationship between the feature and the probability of metastasis, such as diabetes, education, family history, and marital status. For example, the graph for diabetes shows that the probability of metastasis is slightly higher for patients who have diabetes than for patients who do not have diabetes. However, the difference is not very large.
Considering that tumor stage IV is a strong predictor and that modeling tumor cases at earlier stages can have a significant impact on early detection, a separate analysis was conducted on the subset of patients with tumor stages I–III (935 cases, of which 12 had metastasis). The performance of the models was compared with the baseline model based on all stages. Since the performance analysis based on dropping stage IV in the original data was unreliable due to the small sample size of metastasis (12 cases, equal to 1%), only the results of balanced datasets were reported (Table 3). The two approaches, NN and RF, had good performance. The rest of the models did not perform well. Figure 4 demonstrates that the most significant variable is determined by the Gini index. Figure 4A shows that the tumor stage was the most significant factor for predicting metastasis. Subsequently, the number of involved lymph nodes, treatment type, BMI, and tumor size played key roles in predicting metastasis. Figure 4B demonstrates that tumor grade, tumor size, and tumor stage are the most important factors.
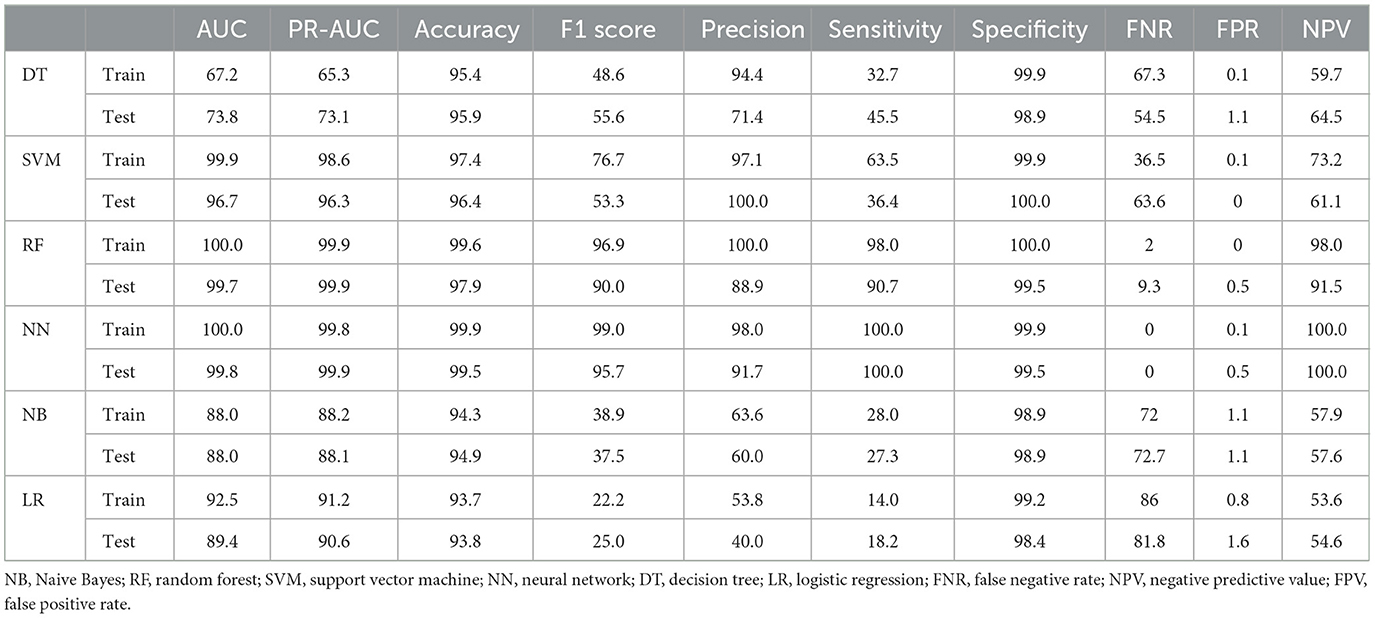
Table 3. Performance criteria for ML methods in predicting metastasis in the balanced dataset of patients with tumor stages I–III.
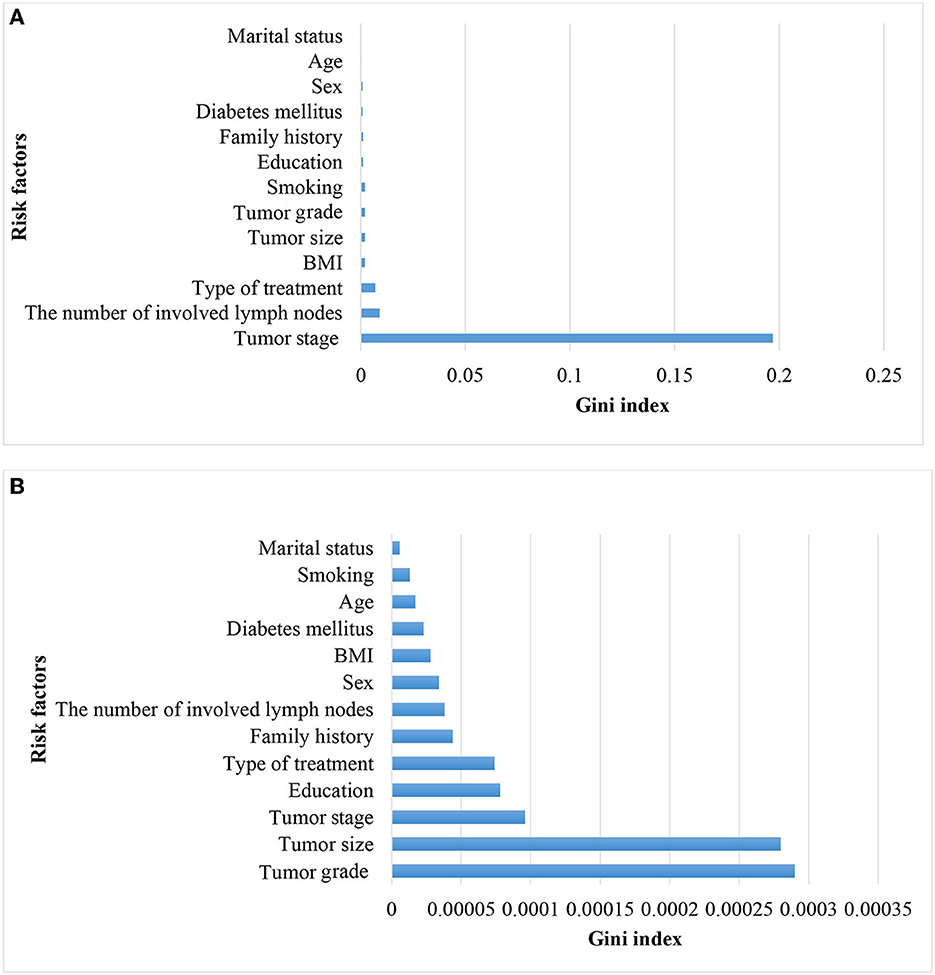
Figure 4. Variable importance for predicting metastasis in patients with CRC in all stages (A) and without CRC in stage IV (B).
Discussion
In the present study, ML models were applied to predict metastasis in CRC patients. The clinical efficacy of our models was determined through ROC curve analysis and other indices, including sensitivity, specificity, and precision. Classifier performance was assessed using the six ML-based approaches. In addition, to focus on modeling tumor cases at earlier stages, which is important for early detection, a separate analysis was conducted for the subset of patients with tumor stages I–III. However, the results of these stages may be unreliable because the number of metastasis samples was very small in these patients.
In our study, while all models exhibited acceptable performance, the NN and RF models demonstrated greater predictive efficiency than the others. A number of ML-based modeling techniques have been suggested for the CRC dataset. Alternative studies have applied DT, SVM, NN, RF, and LR (Cueto-López et al., 2019; Boyne et al., 2020; Nartowt et al., 2020; Achilonu et al., 2021). A study investigated the prediction of tumor staging in colon cancer patients using TNM staging (tumor, node, and metastasis) (Gupta et al., 2019). In this study, ML techniques, such as RF, LR, SVM, NN, k-nearest neighbor (KNN), and adaptive boosting, were applied based on grouping tumor aggression score (TAS) into two categories (>9.8 and <9.8). They concluded that, when tumor size alone was regarded as a prognostic factor, the RF model outperformed other approaches with an accuracy of 84% and 74% in the training and test sets, respectively. In our study, we performed six ML-based approaches using the CRC data, and we found that NN and RF outperformed other models. The RF model was particularly compatible with our study. Moreover, NN and RF presented the highest sensitivity; furthermore, NN and DT showed the highest values in specificity. In their study, both the tumor stage and the number of involved lymph nodes are regarded as the most significant factors. However, the tumor stage was an essential variable, which is consistent with our study.
In 2020, Boyne et al. predicted early discontinuation of adjuvant chemotherapy among individuals aged >17 years with colon cancer patients at a high stage using the LR and RF models (Boyne et al., 2020). Their results revealed that the time from surgery to chemotherapy initiation and the distance from the treatment facility seemed to be the most considerable predictor factors. They concluded that the RF algorithm may help predict early discontinuation of chemotherapy among stage III colon cancer patients. In our study, the NN and RF models were of primary and secondary importance. The primary outcome of their study was chemotherapy discontinuation, defined as a receipt of <5 months and more than 5 months, while metastasis was the dependent variable in our study. In their study, RF was considered a better model than the LR method, while all ML-based approaches exhibited ideal performances in our study.
An investigation was conducted in South Africa using LR, NB, C5.0, RF, SVM, and ANN algorithms for predictive analytics of recurrence and survival outcomes in CRC patients (Achilonu et al., 2021). The analysis considered three datasets, including simulated, recurrent, and survival data. Significant variables in all models were examined and compared using the AUC, which evaluated the discriminatory power of predictive models. This assessment was supported by a threshold (accuracy) metric. Their results demonstrated that all models had the AUC values >80%; however, the ANN model was considered a better method with an AUC of approximately 100%, which was compatible with our study. Nevertheless, an inconsistency arose in the results of the African study, where histology and CRC complications were prioritized in six methods, while in our study, the tumor stage emerged as the primary candidate.
A survey was carried out on the Indonesian population suffering from CRC in four hospitals from 2012 to 2015 (Anuraga and Fernanda, 2019). The predictor factors included the comorbidity, the tumor stage, age, treatment type, cancer location, gender, and metastasis in CRC patients. In this survey, the RF algorithm was employed in data classification, utilizing tree merging through training on sample data. Furthermore, the accuracy of these models was assessed based on the classification value using the AUC. In addition, the most essential variables for the survival of CRC patients were the metastasis history, cancer location, and gender. In our investigation, the outcome variable was metastatic history, whereas the survival of CRC patients served as the dependent variable in the Indonesian study. Moreover, in their study, both the tumor stage and age were of less importance, with the tumor stage being consistent with our survey.
There are some limitations to our study: This study was based on a single tertiary care facility in Iran, which may limit the generalizability of the results to other populations and settings. In addition, the other potential predictors of metastasis, such as the molecular markers, tumor microenvironment, and treatment response, were not considered due to the lack of data availability. The study used a binary outcome of metastasis, which may not capture the complexity of the metastatic process and its clinical implications.
Conclusion
The results of this study indicated that the NN and RF methodscould be the best among ML-based approaches. In addition, in the RF method, the most important variables were the tumor stage, the number of involved lymph nodes, treatment type, and BMI. In a separate analysis of patients with tumor stages I–III, the performance of the NN and RF models was acceptable, with tumor grade, tumor size, and tumor stage identified as the most important factors. However, the results of modeling of I–III stages should be used with caution because the number of metastasis samples was very small.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human data were approved by the Ethics Committee of Iran University of Medical Sciences (IR.IUMS.REC.1400.459). The Ethics Committee waived the requirement for written informed consent.
Author contributions
RT: Investigation, Software, Writing – original draft. CC-M: Supervision, Project administration, Writing – review & editing. AA: Conceptualization, Validation, Writing – review & editing. AT: Formal analysis, Investigation, Writing – review & editing. NB: Formal analysis, Project administration, Software, Supervision, Writing – review & editing. MP: Data curation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors thank Iran University of Medical Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AUC, Area under the curve; BMI, Body mass index; CRC, Colorectal cancer; CV, Cross-validation; DT, Decision tree; LR, Logistic regression; ML, Machine learning; NB, Naive Bayes; NN, Neural network; PR-AUC, Precision–Recall area under the curve; RF, Random forest; ROC, Receiver operating characteristic curve; SVM, Support vector machine.
References
Achilonu, O. J., Fabian, J., Bebington, B., Singh, E., Eijkemans, M. J. C., Musenge, E., et al. (2021). Predicting colorectal cancer recurrence and patient survival using supervised machine learning approach: a South African population-based study. Front. Public Health. 9:694306. doi: 10.3389/fpubh.2021.778749
Anuraga, G., and Fernanda, J. W. (2019). “Random forest prognostic factor in colorectal cancer,” in Journal of Physics: Conference Series (IOP Publishing), 012098. doi: 10.1088/1742-6596/1217/1/012098
Borumandnia, N., Doosti, H., Jalali, A., Khodakarim, S., Charati, J. Y., Pourhoseingholi, M. A., et al. (2021). Nomogram to predict the overall survival of colorectal cancer patients: a multicenter national study. Int. J. Environ. Res. Public Health 18:7734. doi: 10.3390/ijerph18157734
Boyne, D. J., Brenner, D. R., Sajobi, T. T., Hilsden, R. J., Yusuf, D., Xu, Y., et al. (2020). Development of a model for predicting early discontinuation of adjuvant chemotherapy in stage III colon cancer. JCO Clin. Cancer Inform. 4, 972–984. doi: 10.1200/CCI.20.00065
Cueto-López, N., García-Ordás, M. T., Dávila-Batista, V., Moreno, V., Aragonés, N., Alaiz-Rodríguez, R. A., et al. (2019). A comparative study on feature selection for a risk prediction model for colorectal cancer. Comput. Methods Progr. Biomed. 177, 219–229. doi: 10.1016/j.cmpb.2019.06.001
Ferlay, J., Colombet, M., Soerjomataram, I., Mathers, C., Parkin, D. M., Piñeros, M., et al. (2019). Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 144, 1941–1953. doi: 10.1002/ijc.31937
Greener, J. G., Kandathil, S. M., and Moffat, L. (2022). A guide to machine learning for biologists. Nat. Rev. Molec. Cell Biol. 23, 40–55. doi: 10.1038/s41580-021-00407-0
Gupta, P., Chiang, S. F., Sahoo, P. K., Mohapatra, S. K., You, J. F., Onthoni, D. D., et al. (2019). Prediction of colon cancer stages and survival period with machine learning approach. Cancers 11:2007. doi: 10.3390/cancers11122007
Kourou, K., Exarchos, T. P., Exarchos, K. P., and Karamouzis, M. V. (2015). Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 13, 8–17. doi: 10.1016/j.csbj.2014.11.005
Manilich, E. A., Kiran, R. P., Radivoyevitch, T., Lavery, I., Fazio, V. W., Remzi, F. H., et al. (2011). A novel data-driven prognostic model for staging of colorectal cancer. J. Am. Coll. Surg. 213, 579–588.e2. doi: 10.1016/j.jamcollsurg.2011.08.006
Nartowt, B. J., Hart, G. R., Muhammad, W., Liang, Y., Stark, G. F., Deng, J., et al. (2020). Robust machine learning for colorectal cancer risk prediction and stratification. Front. Big Data 3:6. doi: 10.3389/fdata.2020.00006
Nicol,ò, C., Périer, C., Prague, M., Bellera, C., MacGrogan, G., Saut, O., et al. (2020). Machine learning and mechanistic modeling for prediction of metastatic relapse in early-stage breast cancer. JCO Clin. Cancer Inform. 4, 259–274. doi: 10.1200/CCI.19.00133
Siegel, R. L., Miller, K. D., and Jemal, A. (2016). Cancer statistics, 2016. CA. 66, 7–30. doi: 10.3322/caac.21332
Talebi, A., Akbari, A., Mobini, G. R., Ashtari, S., and Pourhoseingholi, M. A. (2019). Biological and clinical relevance of long non-coding RNA PCAT-1 in cancer, a systematic review and meta-analysis. Asian Pacific J. Cancer Prev. 20:667. doi: 10.31557/APJCP.2019.20.3.667
Talebi, A., Celis-Morales, C. A., Borumandnia, N., Abbasi, S., Pourhoseingholi, M. A., Akbari, A., et al. (2023). Predicting metastasis in gastric cancer patients: machine learning-based approaches. Sci. Rep. 13:4163. doi: 10.1038/s41598-023-31272-w
Talebi, A., Mohammadnejad, A., Akbari, A., Pourhoseingholi, M. A., Doosti, H., Moghimi-Dehkordi, B., et al. (2020). Survival analysis in gastric cancer: a multi-center study among Iranian patients. BMC Surg. 20, 1–8. doi: 10.1186/s12893-020-00816-6
Tapak, L., Shirmohammadi-Khorram, N., Amini, P., Alafchi, B., Hamidi, O., and Poorolajal, J. (2019). Prediction of survival and metastasis in breast cancer patients using machine learning classifiers. Clin. Epidemiol. Global Health 7, 293–299. doi: 10.1016/j.cegh.2018.10.003
Wen, R., Zheng, K., Zhang, Q., Zhou, L., Liu, Q., Yu, G., et al. (2021). Machine learning-based random forest predicts anastomotic leakage after anterior resection for rectal cancer. J. Gastrointest. Oncol. 12:921. doi: 10.21037/jgo-20-436
Wieszczy, P., Kaminski, M. F., Franczyk, R., Loberg, M., Kobiela, J., Rupinska, M., et al. (2020). Colorectal cancer incidence and mortality after removal of adenomas during screening colonoscopies. Gastroenterology 158, 875–883. doi: 10.1053/j.gastro.2019.09.011
Keywords: colorectal cancer, machine learning, metastasis, model performance and validation, balance data
Citation: Talebi R, Celis-Morales CA, Akbari A, Talebi A, Borumandnia N and Pourhoseingholi MA (2024) Machine learning-based classifiers to predict metastasis in colorectal cancer patients. Front. Artif. Intell. 7:1285037. doi: 10.3389/frai.2024.1285037
Received: 29 August 2023; Accepted: 03 January 2024;
Published: 24 January 2024.
Edited by:
Junhua Ding, University of North Texas, United StatesReviewed by:
Shailesh Tripathi, University of Applied Sciences Upper Austria, AustriaLeili Tapak, Hamadan University of Medical Sciences, Iran
Copyright © 2024 Talebi, Celis-Morales, Akbari, Talebi, Borumandnia and Pourhoseingholi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nasrin Borumandnia, Ym9ydW1hbmQubkBnbWFpbC5jb20=
 Raheleh Talebi1
Raheleh Talebi1 Carlos A. Celis-Morales
Carlos A. Celis-Morales Abolfazl Akbari
Abolfazl Akbari Atefeh Talebi
Atefeh Talebi Nasrin Borumandnia
Nasrin Borumandnia Mohamad Amin Pourhoseingholi
Mohamad Amin Pourhoseingholi