- 1Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing, China
Male sterility (MS) provides a useful breeding tool to harness hybrid vigor for hybrid seed production. It is necessary to generate new male sterile mutant lines for the development of hybrid seed production technology. The CRISPR/Cas9 technology is well suited for targeting genomes to generate male sterile mutants. In this study, we artificially synthesized Streptococcus pyogenes Cas9 gene with biased codons of maize. A CRISPR/Cas9 vector targeting the MS8 gene of maize was constructed and transformed into maize using an Agrobacterium-mediated method, and eight T0 independent transgenic lines were generated. Sequencing results showed that MS8 genes in these T0 transgenic lines were not mutated. However, we detected mutations in the MS8 gene in F1 and F2 progenies of the transgenic line H17. A potential off-target site sequence which had a single nucleotide that was different from the target was also mutated in the F2 progeny of the transgenic line H17. Mutation in the MS8 gene and the male sterile phenotype could be stably inherited by the next generation in a Mendelian fashion. Transgene-free ms8 male sterile plants were obtained by screening the F2 generation of male sterile plants, and the MS phenotype could be introduced into other elite inbred lines for hybrid production.
Introduction
Male sterility (MS) provides a useful breeding tool to harness hybrid vigor for hybrid seed production, saving labor, and time. MS can be classified into genic male sterility (GMS) and cytoplasmic male sterility (CMS), based on the inheritance pattern. CMS is usually caused by the mutation of a mitochondrial gene that leads to the defective function of plant mitochondria. According to the molecular mechanism of MS, CMS is classified into three types: CMS-T, CMS-C, and CMS-S (Kim and Zhang, 2018). CMS has been widely used in hybrid seed production in three-line systems. However, the application of CMS is limited because of few restorer line resources, instability of the CMS phenotype, and disease susceptibility of CMS-based hybrids (Weider et al., 2009).
Genic male sterile plants are usually generated by mutations of some nuclear gene, and these plants can be used for hybrid seed production in two-line systems. GMS is a more attractive alternative for hybrid breeding than CMS. In 1921, the first maize GMS mutant was identified (Eyster, 1921). Currently, more than 40 maize ms mutants have been reported. Male sterility 44 (MS44) gene was identified to encode a lipid transfer protein that is mainly expressed in the tapetum, and ms44 is a dominant mutant of the gene (Fox et al., 2017). Male sterility 45 (MS45) encodes a strictosidine synthase which plays an important role in the biosynthesis of terpenoid indole alkaloids, and it is required for the formation of the cell wall of male gametophytes (Cigan et al., 2001). Male sterility 32 (MS32) encodes a basic helix-loop-helix (bHLH) transcription factor that regulates the division and differentiation of anther cells (Moon et al., 2013). Male sterility 26 (MS26) encodes a cytochrome P450 monooxygenase, and it plays a role in fatty acid metabolism (Djukanovic et al., 2013). Male sterility 7 (MS7) was cloned by map-based cloning, and it was identified that the gene encodes a plant homeodomain finger transcription factor. The mutation of the MS7 gene led to abnormal programed cell death (PCD) of tapetal cells (Zhang et al., 2018). Male Sterility 8 (MS8) encodes a putative ß-1,3-galactosyltransferase, and it mainly affects the meiotic stage of anther development (Wang et al., 2013). In the previous report, it was shown that the ms8 mutant plants have fewer branches, that they do not exert anthers, and that they have no viable pollen. The anthers of the ms8 mutant plants had more and shorter epidermal cells, while they also had less and larger tapetal cells (Wang et al., 2010). The ms45 mutant has been successfully used in a hybrid production platform called the seed production technology (SPT) (Wu et al., 2015). A multi-control sterility system based on the maize ms7 mutant has also been developed (Zhang et al., 2018).
Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has been used as an efficient genome editing tool. The CRISPR/Cas9 technology is easy to use, and it almost replaces the transcription activator-like effector nuclease (TALEN) technology and the zinc finger nuclease (ZFN) technology. The CRISPR/Cas9 system has been applied for gene targeting in various species, including rice, wheat, cotton, Arabidopsis, tobacco, soybean, and barley (Li et al., 2013; Shan et al., 2013; Jacobs et al., 2015; Gao et al., 2017; Kapusi et al., 2017). Several studies have reported the use of CRISPR/Cas9 in gene targeting in maize (Liang et al., 2014; Svitashev et al., 2015; Feng et al., 2016; Zhu et al., 2016; Shi et al., 2017).
Maize male sterile mutants can also be generated by targeted mutation of candidate genes. Novel ms26 male sterile lines were generated by targeted mutagenesis of MS26 using the redesigned I-CreI homing endonuclease or the CRISPR/Cas9 (Djukanovic et al., 2013;Svitashev et al., 2015). Novel maize ms45 mutants have also been produced by targeted mutagenesis of the MS45 gene (Svitashev et al., 2015). Mutation of the ZmTMS5 gene using the CRISPR/Cas9 technology has generated maize tms5 male sterile mutants that are thermosensitive (Li et al., 2017). In this study, we edited the maize MS8 gene using the CRISPR/Cas9 system, and we obtained novel transgene-free male sterile ms8 lines of maize. These mutant lines are valuable resources for maize hybrid seed production.
Materials and Methods
Construction of the sgRNA-Cas9 Expression Vector
The codons of Streptococcus pyogenes Cas9 (SpCas9) gene were optimized using maize biased codons (Liu and Scott, 2009). The codon-optimized gene was attached with two nuclear localization sequences (NLSs), MSERKRREKL and MISESLRKAIGKR, at the N-terminal and C-terminal ends, respectively. The Cas9 gene was synthesized and cloned into the pUC57 vector by Sangon Biotech (Shanghai, China) to generate the pUC57-Cas9 vector. Using pUC57-Cas9 plasmid as the template, the Cas9 fragment was amplified by PCR with the BsmBI and NcoI sites added to the N-terminal and with the BstEII site added to the C-terminal. The amplified Cas9 fragment was digested with BsmBI and BstEII, and it was inserted into the pCAMBIA3301 plasmid at the NcoI and BstEII sites to generate the pCAMBIA3301-Cas9 vector.
Maize U3 promoter sequence was also synthesized at Sangon Biotech (Shanghai, China), and it was ligated into the pUC57 vector to generate the pUC57-ZmU3 vector. The sequences of Cas9 and the single guide RNA (sgRNA) construct were shown in Supplementary File S1.
A sgRNA targeting the second exon of MS8 (Zm00001d012234) was designed as 5′-GCTGTCCGGGAAGGCCGTCGCGG-3′. Primers 5′-AGCAGCTGTCCGGGAAGGCCGTCG-3′ and 5′-AAACCGACGGCCTTCCCGGACAGC-3′ were synthesized at Sangon Biotech (Shanghai, China). The two primers were mixed and heated at 95°C for 5 min, annealed at room temperature, and then cloned into the pUC57-ZmU3 vector which was digested with BsaI to construct the pUC57-ZmU3-MS8ex2 vector. The pUC57-ZmU3-MS8ex2 vector was digested with BamHI and HindIII, and the ZmU3-MS8ex2 fragment was purified and inserted into the pCAMBIA3301-Cas9 plasmid to construct the pCAMBIA3301-Cas9-MS8ex2 transformation vector (Figure 1).
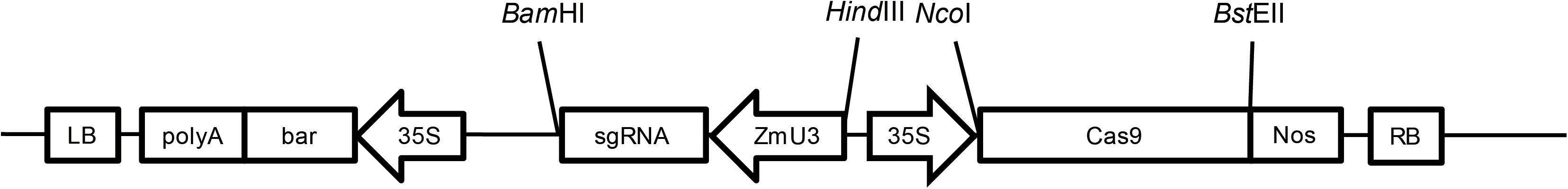
FIGURE 1. The T-DNA region of the pCAMBIA3301-Cas9-MS8ex2 plasmid. The codon-optimized Streptococcus pyogenes Cas9 (SpCas9) gene was inserted into the pCAMBIA3301 plasmid at NcoI and BstEII sites after the 35S promoter. The ZmU3::sgRNA was inserted into the BamHI and HindIII sites. LB, T-DNA left border; polyA, 35S polyA terminator; bar, bar gene; 35S, CaMV 35S promoter; sgRNA, single guide RNA; ZmU3, Zea mays U3 promoter; Cas9, Streptococcus pyogenes Cas9 gene; Nos, Nos terminator; RB, T-DNA right border; BamHI, HindIII, NcoI, BstEII are restriction enzyme sites.
Agrobacterium-Mediated Maize Transformation
The pCAMBIA3301-Cas9-MS8ex2 vector was transformed into the Agrobacterium tumefaciens strain EHA105. Immature embryos from maize hybrid HiII were transformed according to the described method (Zhang et al., 2013) with some modifications. Immature embryos about the size of 1.2 mm were isolated and suspended in liquid infection medium (Murashige and Skoog basal medium, 68.5 g L-1 sucrose, 36.0 g L-1 glucose, 100 μM acetosyringone, pH 5.2). The embryos were heated at 45°C for 3 min, and then transferred to an A. tumefaciens suspension and incubated for 5 min. After inoculation, the embryos were transferred to solid cocultivation medium (half strength Murashige and Skoog basal medium, 20 g L-1 sucrose, 10 g L-1 glucose, 0.85 mg L-1 silver nitrate, 100 μM acetosyringone, 1.22 mg L-1 CuSO4, 8 g L-1 agar, pH 5.8) and were incubated in the dark at 23°C for 3 days. The embryos were then transferred onto resting medium (Murashige and Skoog basal medium, 20 g L-1 sucrose, 10 g L-1 glucose, 1.5 mg L-1 2,4-Dichlorophenoxyacetic acid (2,4-D), 0.85 mg L-1 silver nitrate, 250 mg L-1 cefotaxime, 8 g L-1 agar, pH 5.8) and were cultured at 28°C for 7 days. The embryos were moved to a selection medium (Murashige and Skoog basal medium, 20 g L-1 sucrose, 10 g L-1 glucose, 0.7 mg L-1 proline, 0.25 mg L-1 myo-inositol, 1.5 mg L-1 2,4-D, 0.5 mg L-1 6-Benzylaminopurine (6-BA), 1.22 mg L-1 CuSO4, 250 mg L-1 cefotaxime, 1.5 mg L-1 bialaphos, 8 g L-1 agar, pH 5.8) and were maintained for 2 weeks under dim light (10 μmol m-2 s-1) at 28°C. Then, bialaphos was increased to 3 mg L-1 in the selection medium for two rounds of 2-week selection. Resistant calli were placed in Murashige and Skoog medium containing 1 mg L-1 bialaphos for regeneration under fluorescent white light in a 16/8 h light/dark cycle. The regenerated shoots were transferred to Murashige and Skoog rooting medium. Two weeks later, the regenerated T0 seedlings were transferred to soil and were grown in a greenhouse with 16/8 h light/dark cycle at 25–28°C.
Maize Propagation
F1 generation plants were produced by crossing the transgenic line H17 with the maize inbred line Zong31. These F1 plants were self-pollinated to produce the F2 generation. Backcrossing of the MS trait to maize inbred lines was performed by crossing transgene-free male sterile mutant plants ms8-6 or ms8-18 with maize inbred line Zheng58, respectively. The generated F1 plants were then self-pollinated to produce the F2 plants. The F1 and F2 plants were grown in the field in Beijing or Hainan Province, China.
PCR Analysis of the Transgenic Plants
Genomic DNA was extracted from the leaves of transgenic maize plants using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). For determining the mutations in the target gene, fragments flanking the target site were amplified by PCR using the genomic DNA as the template. For the transgenic male fertile F1 generation plants, PCR products were cloned into pEASY®-T1 Simple Cloning vectors (TransGen, Beijing, China) and five randomly selected individual clones were sequenced, or the PCR products were directly sequenced. For the transgenic male sterile F2 generation plants, PCR products were directly sequenced. For the determination of transgene-free male sterile plants, several fragments at the T-DNA region of the pCAMBIA3301-Cas9-MS8ex2 vector were amplified by PCR with the genomic DNA as the template. The PCR products were analyzed by agarose gel electrophoresis. Primers are listed in Supplementary Table S1.
Phenotype Observation
At the flowering stage, the tassels of wild type (WT) and mutants were photographed in the field in Beijing, China. The spikelets were photographed in the laboratory, and the anthers were observed using an Olympus SZX7.
Inheritance Analysis of CRISPR-Mediated ms8 Genes
F2 plants were grown in the field in Beijing, China, and the phenotype of F2 plants were determined at the flowering stage. The numbers of male sterile plants and male fertile plants were recorded and analyzed using a χ2-test in Microsoft Excel.
Results
Knockout of MS8 Gene in the Progeny of Transgenic Maize Plants
After maize transformation, eight independent T0 transgenic lines were generated. These transgenic lines were confirmed by PCR analysis of both Cas9 gene and bar gene (data not shown). To identify whether the MS8 genes in these transgenic lines were mutated, the targeted region of the MS8 gene was amplified and sequenced directly. Unexpectedly, sequencing results showed that none of these T0 transgenic lines were mutated at the MS8 gene. These T0 transgenic plants could not be self-pollinated to generate T1 generation plants in the greenhouse; hence, they were crossed with maize inbred line Zong31 to produce F1 generation, which was then self-pollinated to produce F2 progeny. To determine the mutations in the target site in F1 plants, fragments flanking the target site were amplified by PCR and were directly sequenced. The sequencing chromatograms of all the 12 F1 plants had chaotic peaks after the target site (Supplementary Figure S1), indicating that there were hemizygous mutations in the MS8 gene at the target site in these plants. These PCR products were then cloned into pEASY®-T1 Simple Cloning vectors (TransGen, Beijing, China), and five randomly selected individual clones were sequenced. The sequencing results confirmed that two types of mutations occurred. One was named as ms8-DelG that had a guanine nucleotide deletion, and the other was named as ms8-InA that had an adenine nucleotide insertion (Supplementary Figure S1). Both of these two mutation types resulted in frame shift and a non-functional MS8 protein. The genotypes of these F1 plants were shown in Supplementary Table S2.
Among the 23 F2 progeny plants from the ms8-DelG-1 line and the 27 F2 progeny plants from the ms8-DelG-3 line (Supplementary Table S2), nine plants (ms8-1, ms8-2, ms8-3, ms8-4, ms8-5, ms8-6, ms8-7, ms8-8, and ms8-9) and six plants (ms8-19, ms8-20, ms8-21, ms8-22, ms8-23, and ms8-24) showed male sterile phenotype, respectively. Sequencing the ms8 gene in these male sterile plants identified the homozygous mutation in ms8-DelG (Figure 2). Among the 27 F2 progeny plants obtained from line ms8-InA-2 (Supplementary Table S2), nine plants (ms8-10, ms8-11, ms8-12, ms8-13, ms8-14, ms8-15, ms8-16, ms8-17, and ms8-18) showed male sterile phenotype. Sequencing the ms8 gene in these male sterile plants identified the homozygous mutation in ms8-InA (Figure 2). We grew more F2 progeny plants of these three lines to assess the ratio of male fertile and male sterile plants, and we found a 3:1 segregation between fertile and sterile plants (Table 1). This result suggested that the male sterile phenotype was inherited in a Mendelian fashion.
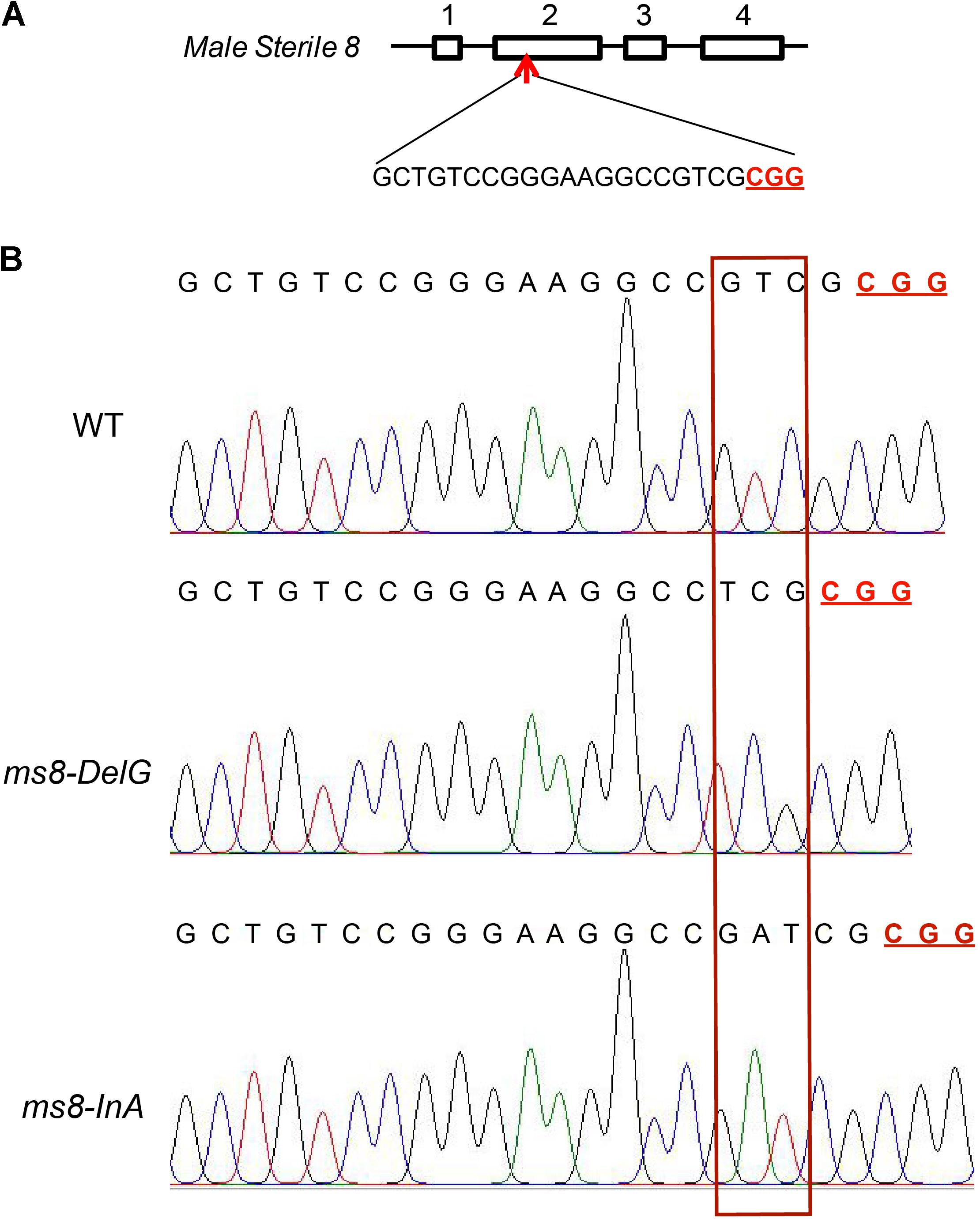
FIGURE 2. Gene-targeting of MS8 gene in male sterile F2 plants. (A) Diagram of the MS8 gene and the target site; four exons were numbered; the PAM motif is underlined and in bold. (B) The sequencing chromatograms of MS8 in maize inbred line B73, ms8-DelG/ms8-DelG, and ms8-InA/ms8-InA plants. T0 transgenic line H17 plant was crossed with maize inbred line Zong31 to produce F1 generation, which was then self-pollinated to produce F2 progeny. Fragments flanking the target site were amplified by PCR using the genomic DNA of male sterile F2 plants as the template, and the PCR products were directly sequenced.

TABLE 1. Segregation analysis of the F2 generation produced by crossing transgenic T0 line H17 and inbred line Zong31.
Similar to WT plants, new ms8 mutant plants have normal vegetative growth and female fertility. The phenotypes of these male sterile plants were significantly different from that of the WT at the flowering stage. The WT anthers were non-translucent because the anthers were full of pollen grains (Figures 3A,D,G), whereas none of the mutant plants had exerted anthers (Figures 3B,C). Mature mutant anthers were empty and half-translucent when viewed on a light table, without any visible pollen grains (Figures 3E,F,H,I). At the late stage, the mutant anthers began senescing and shrinking; meanwhile, the mutant tassel became white and eventually withered.

FIGURE 3. The phenotype of F2 mutant and wild type plants. The phenotype of tassels, spikelets, and anthers of WT (A,D,G), ms8-DelG/ms8-DelG (B,E,H), and ms8-InA/ms8-InA (C,F,I). T0 transgenic line H17 plant was crossed with maize inbred line Zong31 to produce F1 generation, which was then self-pollinated to produce F2 progeny. At the flowering stage, the WT tassel had anthers, and the anthers were full of pollen, whereas tassels from the two mutants did not exert anthers, and anthers were empty without any pollen. Bar = 1 cm in D–F; bar = 1 mm in G–I.
Generation of Transgene-Free Male Sterile Plants
Because the MS phenotype was inherited in a Mendelian fashion, we speculated that transgene-free male sterile plants could be obtained in the progeny. To obtain transgene-free ms8 male sterile plants, we used PCR to screen 15 F2 male sterile plants from the ms8-DelG-1 and ms8-DelG-3 lines. We also screened nine F2 male sterile plants from the ms8-InA-2 line. Six pairs of primers were designed to amplify Cas9, sgRNA, and 35S::bar fragments (Figure 4A). Results showed that transgene fragments existed in 11 F2 mutant plants from the ms8-DelG-1 and ms8-DelG-3 lines, and seven F2 mutant plants from the ms8-InA-2 line, respectively. However, the transgene fragment was eliminated in four F2 male sterile plants from the ms8-DelG-1 and ms8-DelG-3 lines (ms8-6, ms8-20, ms8-22, and ms8-24) and in two F2 male sterile plants from the ms8-InA-2 line (ms8-14 and ms8-18) (Figure 4B). These results suggested that mutations of the MS8 gene were fixed in these transgene-free plants.
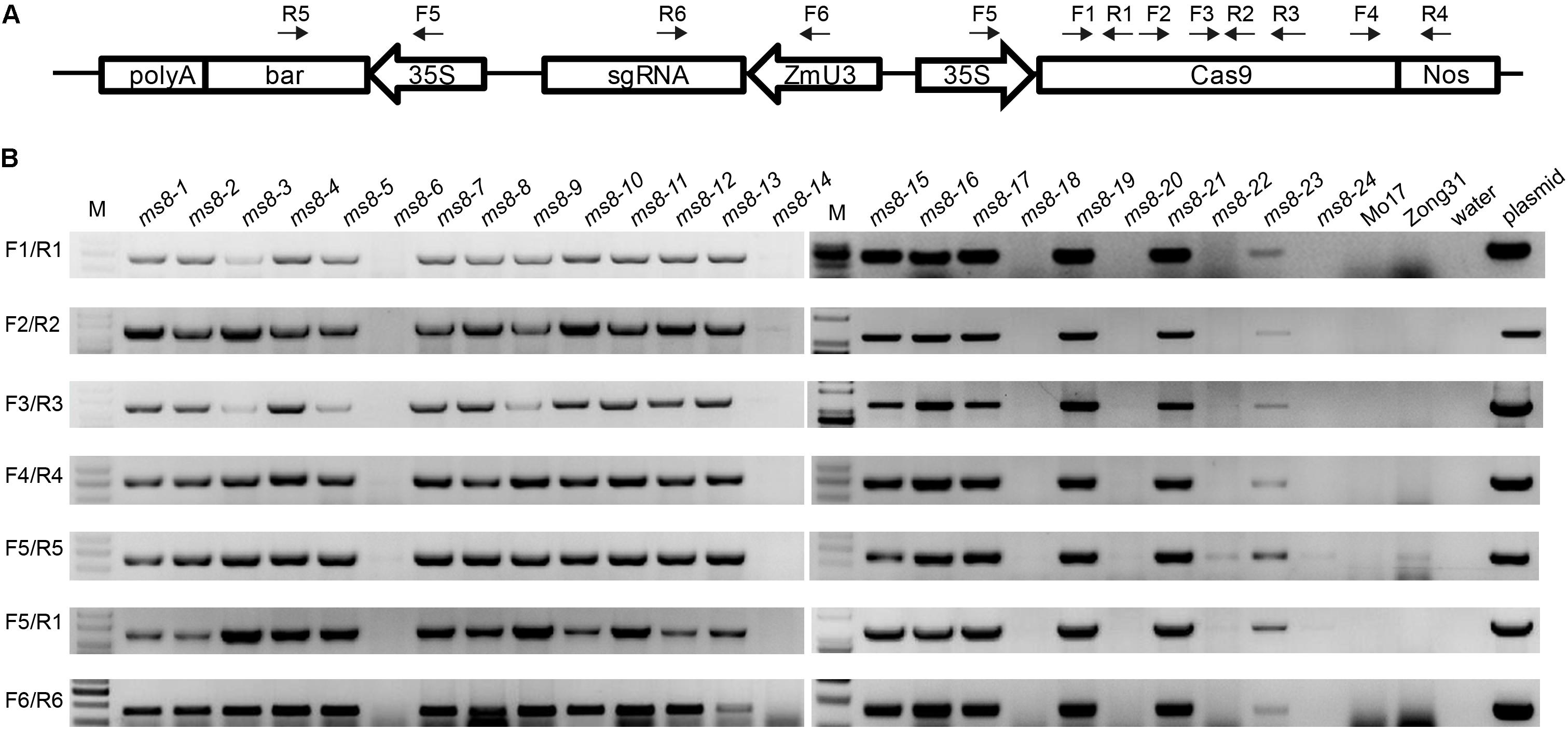
FIGURE 4. PCR screening of transgene-free male sterile F2 plants. (A) Schematic of the T-DNA region of the pCAMBIA3301-Cas9-MS8ex2 vector and the location of the primers. (B) Agarose gel electrophoresis results of seven fragments of T-DNA. M, DNA molecular weight marker; ms8-1–ms8-9: the male sterile F2 plants from the ms8-DelG-1 line; ms8-10–ms8-18: the male sterile F2 plants from the ms8-InA-2 line; ms8-19–ms8-24: the male sterile F2 plants from the ms8-DelG-3 line; Mo17: maize inbred line Mo17; Zong31: maize inbred line Zong31.
To further investigate the inheritability of MS in these transgene-free mutants, we crossed two transgene-free male sterile plants ms8-6 and ms8-18, which belong to the ms8-DelG-1 and ms8-InA-2 lines, with maize elite inbred line Zheng58. All of the F1 progeny plants were male fertile, whereas the F2 population displayed a 3:1 segregation ratio between male fertile and male sterile plants (Table 2). These results suggested that the MS phenotype could be introduced to other elite inbred lines for hybrid production.
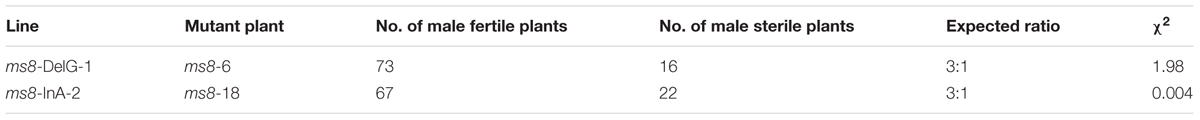
TABLE 2. Segregation analysis of the F2 generation produced by crossing transgene-free male sterile plants and inbred line Zheng58.
Evaluation of Off-Target Mutations in Transgene-Free Male Sterile Mutants
Off-target mutations using the CRISPR/Cas9 system have been reported in many crops, including maize (Feng et al., 2016), rice (Endo et al., 2015), and soybean (Jacobs et al., 2015). To evaluate the possibility of off-target mutations induced by CRISPR/Cas9 in this study, we searched the potential off-target sites in the CRISPR-P website1. Nine potential off-target sites were found to have 95 or 85% identity with the target sequence. The fragments flanking four potential off-target sites were amplified from male sterile mutants and sequenced (Table 3). Sequencing results showed that mutations occurred in the potential off-target site sequence which had a single nucleotide that was different from the target. However, mutations did not occur in the sequences which had three nucleotides that were different from the target (Table 3). For the potential off-target site sequences which had a single nucleotide that was different from the target, a cytosine nucleotide deletion allele was identified in some male sterile plants, but the genotypes were not associated with the male sterile phenotype (Supplementary Table S3).
Discussion
Compared with CMS, GMS plant materials are much more useful for the hybrid SPT. Environment-sensitive GMS has been explored in two-line hybrid production system for wheat and rice (Whitford et al., 2013; Huang et al., 2014). Using GMS mutant ms45, DuPont Pioneer developed a hybrid production platform named SPT (Wu et al., 2015). Zhang et al. (2018) also developed a multi-control sterility system based on the ms7 mutant. Expanding male sterile germplasm resources is necessary for developing new SPT technology. The MS phenotype is usually controlled by the recessive mutation of nuclear gene; therefore, it is possible to generate male-sterile mutants using the CRISPR/Cas9 system. Several MS genes have been identified to control the development of pollen or anther. In this study, we chose male sterility 8 (MS8) as the targeted gene and generated novel ms8 mutant lines using the CRISPR/Cas9 system. The MS phenotype of the novel ms8 lines generated in this study could be introduced to other elite inbred lines for hybrid production, which is essential for the use of these mutants in producing hybrids. The new ms8 lines generated in this study showed a male sterile phenotype similar to the reported natural ms8 mutant in which the Mu transposon was inserted in the MS8 gene (Wang et al., 2013).
If the CRISPR/Cas9 system works well, then there should be high editing efficiency in T0 generation plants. In this study, we did not observe mutations in the MS8 gene in the T0 plants. T0 plants were crossed with maize inbred line Zong31 to generate F1 plants, and the editing of MS8 gene in F1 plants was observed. The main reason for the low mutation efficiency of CRISPR/Cas9 system might be that the sgRNA is not stable and has poor targeting efficiency. It has been shown that the low activity of 35S promoter in germline cells would lead to low editing efficiency (Fauser et al., 2014). Using meiotic promoter to drive the expression of Cas9 could increase the targeted mutagenesis efficiency (Eid et al., 2016; Mao et al., 2016). In this study, the Cas9 gene was driven by the 35S promoter and its activity might be low. To localize the Cas9 protein into the nucleus, we fused the amino acids MSERKRREKL and MISESLRKAICKR (Shieh et al., 1993) to the N- and C-terminal ends of the Cas9 protein. We speculate that these nuclear location sequences (NLS) might not completely transport the target Cas9 protein to the nucleus, which might be another reason for the low mutation efficiency.
Off-target events often occurred during the gene targeting process using the CRISPR/Cas9 system. To reduce off-target editing, several strategies such as SpCas9-HF1 variants (Kleinstiver et al., 2016) and CRISPR/Cas9 nickase have been used (Shen et al., 2014). In our study, off-target mutations occurred in sequence which had a single nucleotide that was different from the target. However, there were no mutations in the sequences which had three nucleotides that were different from the target sequence. In some cases, off-target mutations might lead to the defective growth of plants. In this study, the off-target mutations in the ms8 mutant did not lead to a negative effect on the mutant plants. Off-target mutations could also be eliminated by using the breeding methods of backcrossing or outcrossing.
Globally, there is a strict regulatory framework for transgenic crops. Using preassembled CRISPR/Cas9 ribonucleoproteins (RNPs) could generate transgene-free plants with edited genomes (Woo et al., 2015). CRISPR/Cas9 RNPs have also been successfully used to produce transgene-free genome-edited maize (Svitashev et al., 2016) and wheat (Liang et al., 2017). For the genome-edited crops, transgenes and off-targeted genes could also be eliminated by outcrossing or backcrossing. In general, the regulations for genome-edited crops without transgenes should be different from the traditional transgenic crops (Huang et al., 2016). In our study, transgenes were eliminated by backcrossing and transgene-free male sterile mutants were generated. These transgene-free male sterile mutants could be used for maize hybrid production systems without the limitation of regulatory frameworks for transgenic organisms. For application purposes, the MS phenotype of transgene-free mutants should be introduced into different elite maize lines by backcrossing.
Conclusion
In this study, a CRISPR/Cas9 vector targeting the MS8 gene of maize was constructed, and it was transformed into maize plants using an Agrobacterium-mediated method; consequently, we detected mutations in the MS8 gene in F1 and F2 progeny plants. Mutations in the MS8 gene and the male sterile phenotype could be stably inherited by the next generation in a Mendelian fashion. Transgene-free ms8 male sterile plants were obtained, and the MS phenotype could be introduced into other elite inbred lines for hybrid production.
Author Contributions
RC, GW, and YJL designed the research and wrote the article. RC, QX, YL, JZ, and DR performed the research and analyzed the data.
Funding
This work was supported by the Fundamental Research Funds for Central Non-Profit of Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (S2018QY07) and the National Major Project for Transgenic Organism Breeding (2016ZX08010-004).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers for the valuable advice to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01180/full#supplementary-material
FIGURE S1 | Gene-targeting of MS8 gene in F1 plants produced by crossing T0 transgenic line H17 and inbred line Zong31. (A) The sequencing chromatograms of maize inbred line B73; (B,C) the sequencing chromatograms of ms8-DelG/MS8 plant; (D,E) The sequencing chromatograms of ms8-InA/MS8 plant. In (B,D), PCR products were directly sequenced. In (C,E), PCR products were cloned into pEASY®-T1 Simple Cloning vectors (TransGen, Beijing, China) and the randomly selected individual clone was sequenced. The PAM motif is underlined and in bold.
TABLE S1 | Primers used in this study.
TABLE S2 | The genotypes of F1 plants crossing between transgenic line H17 with inbred line Zong31.
TABLE S3 | The genotypes of the off-target site with a single nucleotide difference.
Footnotes
References
Cigan, A. M., Unger, E., Xu, R. J., Kendall, T., and Fox, T. W. (2001). Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex. Plant Reprod. 14, 135–142. doi: 10.1007/s004970100099
Djukanovic, V., Smith, J., Lowe, K., Yang, M., Gao, H., Jones, S., et al. (2013). Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 76, 888–899. doi: 10.1111/tpj.12335
Eid, A., Ali, Z., and Mahfouz, M. M. (2016). High efficiency of targeted mutagenesis in Arabidopsis via meiotic promoter-driven expression of Cas9 endonuclease. Plant Cell Rep. 35, 1555–1558. doi: 10.1007/s00299-016-2000-4
Endo, M., Mikami, M., and Toki, S. (2015). Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 56, 41–47. doi: 10.1093/pcp/pcu154
Eyster, L. (1921). Heritable characters of maize VII. Male sterile. J. Hered. 12, 138–141. doi: 10.1093/jhered/12.3.138
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. doi: 10.1111/tpj.12554
Feng, C., Yuan, J., Wang, R., Liu, Y., Birchler, J. A., and Han, F. (2016). Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics 43, 37–43. doi: 10.1016/j.jgg.2015.10.002
Fox, T., Debruin, J., Haug, C. K., Trimnell, M., Clapp, J., Leonard, A., et al. (2017). A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 15, 942–952. doi: 10.1111/pbi.12689
Gao, W., Long, L., Tian, X., Xu, F., Liu, J., Singh, P. K., et al. (2017). Genome editing in cotton with the CRISPR/Cas9 system. Front. Plant Sci. 8:1364. doi: 10.3389/fpls.2017.01364
Huang, J. Z., Zg, E., Zhang, H. L., and Shu, Q. Y. (2014). Workable male sterility systems for hybrid rice: genetics, biochemistry, molecular biology, and utilization. Rice 7:13. doi: 10.1186/s12284-014-0013-6
Huang, S., Weigel, D., Beachy, R. N., and Li, J. (2016). A proposed regulatory framework for genome-edited crops. Nat. Genet. 48, 109–111. doi: 10.1038/ng.3484
Jacobs, T. B., Lafayette, P. R., Schmitz, R. J., and Parrott, W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16. doi: 10.1186/s12896-015-0131-2
Kapusi, E., Corcuera-Gomez, M., Melnik, S., and Stoger, E. (2017). Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Front. Plant Sci. 8:540. doi: 10.3389/fpls.2017.00540
Kim, Y. J., and Zhang, D. (2018). Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. doi: 10.1016/j.tplants.2017.10.001
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi: 10.1038/nature16526
Li, J., Zhang, H., Si, X., Tian, Y., Chen, K., Liu, J., et al. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genomics 44, 465–468. doi: 10.1016/j.jgg.2017.02.002
Li, J. F., Norville, J. E., Aach, J., Mccormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Liang, Z., Chen, K. L., Li, T. D., Zhang, Y., Wang, Y. P., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8:14261. doi: 10.1038/ncomms14261
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Liu, D., and Scott, M. P. (2009). Design of gene constructs for transgenic maize. Methods Mol. Biol. 526, 3–20. doi: 10.1007/978-1-59745-494-0_1
Mao, Y., Zhang, Z., Feng, Z., Wei, P., Zhang, H., Botella, J. R., et al. (2016). Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 14, 519–532. doi: 10.1111/pbi.12468
Moon, J., Skibbe, D., Timofejeva, L., Wang, C. J. R., Kelliher, T., Kremling, K., et al. (2013). Regulation of cell divisions and differentiation by male sterility32 is required for anther development in maize. Plant J. 76, 592–602. doi: 10.1111/tpj.12318
Murray, M., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shen, B., Zhang, W., Zhang, J., Zhou, J., Wang, J., Chen, L., et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402. doi: 10.1038/nmeth.2857
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shieh, M. W., Wessler, S. R., and Raikhel, N. V. (1993). Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol. 101, 353–361. doi: 10.1104/pp.101.2.353
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., and Mark, A. C. (2016). Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7:13274. doi: 10.1038/ncomms13274
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Wang, D., Oses-Prieto, J. A., Li, K. H., Fernandes, J. F., Burlingame, A. L., and Walbot, V. (2010). The male sterile 8 mutation of maize disrupts the temporal progression of the transcriptome and results in the mis-regulation of metabolic functions. Plant J. 63, 939–951. doi: 10.1111/j.1365-313X.2010.04294.x
Wang, D. X., Skibbe, D., and Walbot, V. (2013). Maize Male sterile 8 (Ms8), a putative beta-1,3-galactosyltransferase, modulates cell division, expansion, and differentiation during early maize anther development. Plant Reprod. 26, 329–338. doi: 10.1007/s00497-013-0230-y
Weider, C., Stamp, P., Christov, N., Foueillassar, X., Camp, K. H., and Munsch, M. (2009). Stability of cytoplasmic male sterility in maize under different environmental conditions. Crop Sci. 49, 77–84. doi: 10.1051/ebr/2010009
Whitford, R., Fleury, D., Reif, J. C., Garcia, M., Okada, T., Korzun, V., et al. (2013). Hybrid breeding in wheat: technologies to improve hybrid wheat seed production. J. Exp. Bot. 64, 5411–5428. doi: 10.1093/jxb/ert333
Woo, J. W., Kim, J., Kwon, S. I., Corvalan, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Wu, Y., Fox, T. W., Trimnell, M. R., Wang, L., Xu, R. J., Cigan, A. M., et al. (2015). Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 14, 1046–1054. doi: 10.1111/pbi.12477
Zhang, D., Wu, S., An, X., Xie, K., Dong, Z., Zhou, Y., et al. (2018). Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 16, 459–471. doi: 10.1111/pbi.12786
Zhang, Y., Liu, Y., Ren, Y., Liu, Y., Liang, G., Song, F., et al. (2013). Overexpression of a novel Cry1Ie gene confers resistance to Cry1Ac-resistant cotton bollworm in transgenic lines of maize. Plant Cell Tissue Organ Cult. 115, 151–158. doi: 10.1007/s11240-013-0348-5
Keywords: maize, CRISPR/Cas9, genome editing, male sterility, transgene-free
Citation: Chen R, Xu Q, Liu Y, Zhang J, Ren D, Wang G and Liu Y (2018) Generation of Transgene-Free Maize Male Sterile Lines Using the CRISPR/Cas9 System. Front. Plant Sci. 9:1180. doi: 10.3389/fpls.2018.01180
Received: 15 February 2018; Accepted: 24 July 2018;
Published: 07 September 2018.
Edited by:
Joachim Hermann Schiemann, Julius Kühn-Institut, GermanyReviewed by:
Eva Stoger, Universität für Bodenkultur Wien, AustriaMahmudul Hassan, Patuakhali Science and Technology University, Bangladesh
Copyright © 2018 Chen, Xu, Liu, Zhang, Ren, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoying Wang, wangguoying@caas.cn Yunjun Liu, liuyunjun@caas.cn
 Rongrong Chen
Rongrong Chen Qilong Xu1
Qilong Xu1 Jiaojiao Zhang
Jiaojiao Zhang Dongtao Ren
Dongtao Ren Guoying Wang
Guoying Wang Yunjun Liu
Yunjun Liu