- 1Institute of Biology and Environmental Sciences, Carl von Ossietzky University of Oldenburg, Oldenburg, Germany
- 2Institute for Chemistry and Biology of the Marine Environment, Carl von Ossietzky University of Oldenburg, Oldenburg, Germany
- 3Department of Anatomy, University of Otago, Dunedin, New Zealand
Climatic changes and anthropogenic pressures affect biodiversity and community composition. These biodiversity shifts are recognized in marine ecosystems, but the underlying processes are barely understood so far. Importantly, human well-being highly relies on oceanic services, which are affected by anthropogenic pressures. Here, we review how interdisciplinary research approaches, with the incorporation of eDNA (environmental DNA) analyses, can help increase the understanding of complex ecosystem processes and dynamics, and how they affect ecosystem services. We discuss marine conservation issues in the light of life cycle aspects and conclude that eDNA can improve our ecological knowledge in some instances, for example, in tracking migration patterns. We also illustrate and discuss the application of eDNA analysis within the context of population genetics, epigenetics, geochemistry and oceanography. Embedded into an interdisciplinary context, eDNA can be exploited by a huge variety of methodological techniques, and can resolve spatio-temporal patterns of diversity, species, or even populations within ecological, evolutionary, and management frameworks.
Introduction
Across the globe, biodiversity declines and extinction rates have accelerated over the last few decades due to climate change and anthropogenic influences (IPBES, 2019). More data about recent biodiversity change is needed to outline the underlying mechanism of these processes (IPBES, 2019). Ecological theory and modeling rely on this data for accurately predicting the future of ecosystem services. With the help of model predictions, stakeholders are able to minimize negative effects. Understanding how communities respond to environmental change is an important aspect of monitoring biodiversity.
Anthropogenic influences are one cause of biological diversity change (Halpern et al., 2008; McGill et al., 2015). Human activities heavily shape marine ecosystems with important consequences for human economies and health (McGill et al., 2015; Culhane et al., 2018). For example, coastal areas can provide a variety of ecosystem services such as flood prevention (e.g., mangroves), food, maintaining water quality, furthering medical discoveries, and decreasing disease vulnerability (Worm et al., 2006; Bernstein and Ludwig, 2008; Mace et al., 2012). Between 1970 and 2007, marine fisheries declined by 38% globally and are projected to decline further (Worm et al., 2009; Hutchings et al., 2010). Human-facilitated carbon release is also causing unprecedented levels of ocean acidification (IPCC, 2014). To know how anthropogenic influences have affected the seascape biodiversity, constant and consistent monitoring is needed (Kavanaugh et al., 2016).
People are willing, more than ever before, to invest in the restoration of nature due to increasing awareness of biodiversity’s benefits (IPBES, 2019). Especially in the marine environment, conservation is becoming more important due to increased fisheries and overfishing. But in the past, protection efforts rarely met the expected outcomes (McClanahan et al., 2006). More research is needed to understand complex ecosystem processes, such as in climatic influences (Cardinale et al., 2012), resource use efficiency (RUE) (Hodapp et al., 2019) or geochemical cycles. To evaluate the conservation success of protecting an area, often just a few key species are monitored (e.g., Edgar et al., 2014) due to limited financial issues. Monitoring changes in species composition and abundance within the context of the biotic community can help to evaluate the success of protection efforts.
In order to extend existing monitoring techniques with highly effective methods, identification of species with molecular methods find its way into monitoring approaches. Hebert et al. (2003) put forth DNA barcoding to identify unknown organism tissues down to species level. This turned out to be a useful tool because DNA barcoding identifies species as well as other methods do (Krishnamurthy and Francis, 2012). Traditionally, barcoding studies classify organisms into molecular operational taxonomic units (MOTUs/OTUs) (e.g., Creer et al., 2010). These MOTUs/OTUs group highly similar barcodes together, collapsing intraspecific sequence variations and estimating interspecific similarity. To assign these MOTUs/OTUs to a certain species, traditional taxonomic classification needs to be used. This integrative taxonomy is highly recommended for research in conservation issues (Krishnamurthy and Francis, 2012). When traditional species identification is challenging or not possible, barcoding can sustain the reliability of results. Notable taxonomic challenges include larval stages and cryptic species, which may be best identified genetically (Ball et al., 2005). Genetic approaches of species identification rely highly on robust databases. Since databases are large and not always well curated, sequence comparison can sometimes lead to ambiguous results. Databases, like BOLD (Barcode of Life1), using integrative taxonomy approaches to ensure robustness of their database. Molecular identification coupled with robust taxonomic identification is increasingly important for building accurate genetic databases. The decline in funding for taxonomy and decrease in specialized taxonomic identification skill makes barcoding a useful tool if properly curated databases can be used. Databases using integrative taxonomy are highly recommended to ensure an accurate assignment of sequence information to species.
A new methodology, the analysis of environmental DNA (eDNA) can help to address gaps in our knowledge about biological community change (Figure 1). eDNA is characterized as a mixture of DNA shed (through skin, mucous, feces, etc.) by many organisms into the environment (soil, water, air) (Bohmann et al., 2014). eDNA can be used to target single species, like invasive or rare species (e.g., Wilcox et al., 2018), but also can be used to investigate whole community compositions when combined with metabarcoding approaches. Cost effectiveness, sensitivity, and non-invasiveness (e.g., Smart et al., 2015, 2016) are just a few benefits of eDNA approaches, helping to increase the understanding of biodiversity in a changing world.
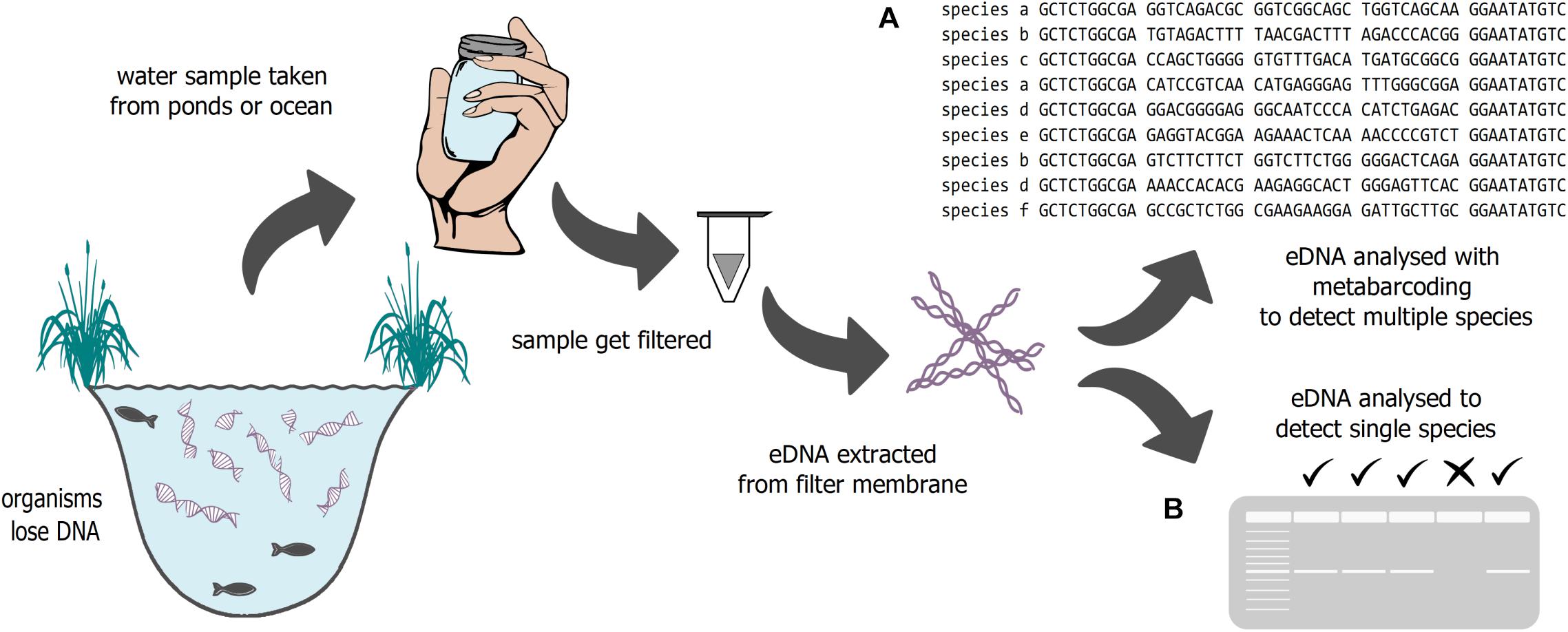
Figure 1. Schematic illustration of eDNA sampling. DNA fragments are filtered out of water and extracted. The fragments can be analyzed either to investigate (A) community structure or (B) the presence or absence of certain species.
Environmental DNA analysis can be used to target one or multiple species (Figure 1). In some cases, single species approaches using eDNA can be very beneficial, for example the use of qPCR for early detection of invasive species (see Section “Ecological Theory – An Overview”), or to estimate relative abundances of species (see Sections “Fisheries Management” and “Rare and Endangered Species”). If only a single species is pertinent to the interest of a study, some evidence suggests species-specific qPCR methods may be slightly more sensitive to detection than multi-species metabarcoding eDNA approaches (Harper et al., 2018; Bylemans et al., 2019). Environmental DNA metabarcoding, which is one of the most common techniques to detect species diversity, can be used to survey multiple taxa on both spatial and temporal scales for bioassessment (Taberlet et al., 2012, 2018; Stoeckle et al., 2017; Pawlowski et al., 2018). This technique uses PCR to amplify informative genetic targets using universal PCR primers, usually in commonly amplified mitochondrial, ribosomal, or nuclear regions. With eDNA metabarcoding, it is possible to identify multiple taxa, usually within a specific clade, such as fish or metazoans (Valentini et al., 2016; Ruppert et al., 2019). A similar approach is Tree of Life (ToL) metabarcoding where eDNA is shotgun sequenced to assess biodiversity at an holistic ecosystem level (Stat et al., 2017).
Dark diversity, biological diversity in an area which is not identified via traditional methods, may be obtained by eDNA methods (Boussarie et al., 2018; Cowart et al., 2018). This can help illuminate a fuller picture of a studied ecosystem. As well, eDNA can improve detection rates of poorly known and difficult to sample organisms (Wilcox et al., 2016). Using eDNA has the advantage of identifying species which are cryptic or taxonomically challenging to identify, e.g., larval stages, compared to more traditional methods such as nets and visual surveys (Thomsen et al., 2012b; Schmelzle and Kinziger, 2016; Yamamoto et al., 2017). Besides identifying an increased or more accurate list of taxa, eDNA can be used for monitoring changes in marine community composition. Already, eDNA has been used to document some community-level shifts in a variety of human-mediated environments (Pawlowski et al., 2014).
Since Ficetola et al. (2008) used eDNA to detect macrofaunal species presence, the use of eDNA has grown exponentially. With increasingly interdisciplinary research, we can see the power of combining singular techniques and methods to obtain a fuller picture of ecological patterns and processes. For example, combining observations of community change with geochemical parameters (e.g., Osterholz et al., 2016) or combining species distribution patterns with hydrodynamics (e.g., Stuckas et al., 2017) has yielded information about how communities are changing. Environmental DNA analysis can also be used in other fields, not just to monitor biodiversity. Implementation of eDNA analysis, for example, in the field of population genetics or toxicology (Zhang, 2019), can offer a huge potential (Adams et al., 2019). The fusion of advanced technologies can result in highly promising tools, helping us save our oceans.
In this review, we focus on the implementation of eDNA-based approaches in an interdisciplinary context for marine management and conservation. We focus on eDNA studies in the marine environment, but also include some freshwater studies to give a broad overview of possible applications. Furthermore, we give an overview on current state of relevant ecological theories and their implementation into marine management and conservation. We do our best to include all relevant publications to displaying the state of these fields, but the rapidly expanding eDNA literature is not necessarily limited to just the cited studies. Here, we highlight studies where eDNA approaches were used in the context of marine monitoring. We also provide an outlook on future possibilities for improving interdisciplinary research approaches.
Marine Management and Ecosystem Functioning
Ecological Theory – An Overview
Marine ecosystems are threatened more than ever from anthropogenic impacts. Increased fishing, environmental pollution, and human-driven climatic change accelerates extinction rates and biodiversity loss in our oceans and around the globe. Stable ecosystems can offer a huge variety of services for humans. The awareness of interrelations between ecosystem services, biodiversity and community composition in stabilizing ecosystems (Figure 2), rose over the last 30 years (Cardinale et al., 2012). The complexity of ecosystem processes requires the gathering of large datasets to understand these interactions. Some of these mechanisms have just recently been discovered, like for example the impact of RUE on a community level (Hodapp et al., 2019) or the stabilizing effect of far-ranging top predators on meta-food webs (Brechtel et al., 2019), and many may still be undocumented. The loss of biodiversity entails a decline in total biomass production (Hillebrand and Cardinale, 2010), which makes fisheries management essential for sustainable future ocean ecosystems. This section gives a simplified overview about the recent state of knowledge in ecosystem theory in relevance to this review.
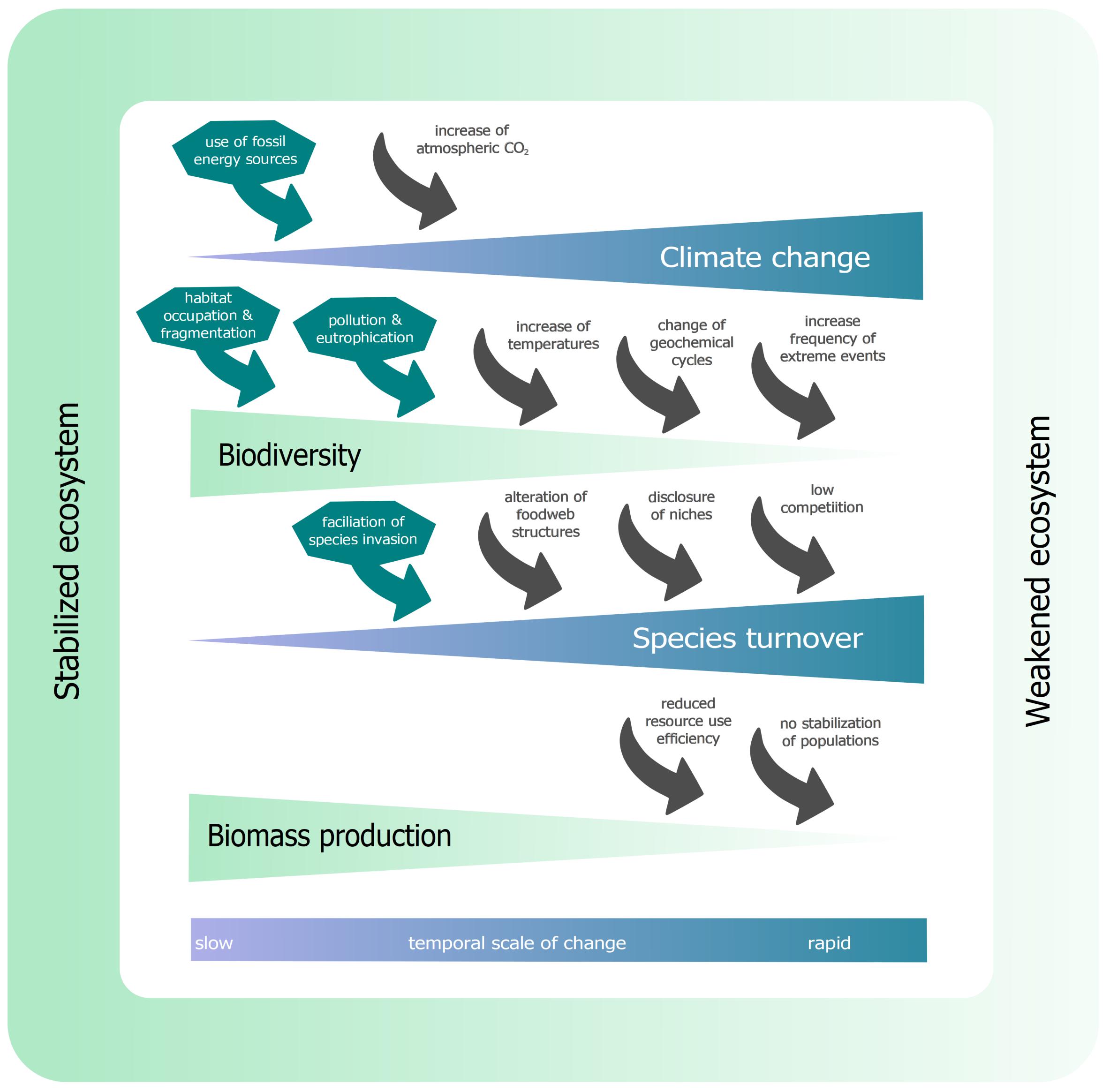
Figure 2. Interactions between climate change, biodiversity and biomass production. Anthropogenic influence (dark green arrows) accelerates climate change, biodiversity loss and species turnover rates from outside the system. Within the ecosystem (gray arrows), changes in climatic conditions force biodiversity loss, which accelerates species turnover rates through the reduction of invasive resistance. These increased turnover rates lead to a reduced biomass production. This figure is a composition of various studies, modeling the effects within ecosystem dynamics, including meta-analysis studies (see Section “Ecological Theory – An Overview”).
The acceleration of climatic changes in the last century is one of the major drivers of biodiversity loss and the reduction of biomass production (IPCC, 2014). The use of fossil energy sources is the most critical anthropogenic driver (dark green arrows, Figure 2) of increased atmospheric CO2 concentrations, beside deforestation (leading to decrease in CO2 binding capacity). The increased atmospheric CO2 concentrations (+ 40% since 1750) led to global temperature increase, in oceans + 0.11°C per decade (1970-2010) (IPCC, 2014).
This temperature increase can lead to intraspecific changes in physiology as, for instance, in juvenile hydroids (Hydractinia echinata) showed increased respiration rates of up to > 50% when exposed to + 3°C (Eder et al., 2018). Additionally, phenology can be affected by temperature increase. For example, plankton blooms (Ceratium fusus) shifted from September (1958-1980) toward July (1981-2002) (Edwards and Richardson, 2004), leading to interspecific phenological mismatches (e.g., Doney et al., 2012; Asch et al., 2019). Increased temperature can also force more frequent extreme weather events (storms, heavy rainfall, heat waves, etc.), which can alter phenology as well (e.g., Jentsch et al., 2009). Increased temperature has the potential to change biogeochemical cycles (Doney et al., 2012), the most famous example here is reduced growth of calcifying organisms, like diatoms or bivalves (Beaugrand et al., 2012). Additionally, geochemical cycles can be altered due to anthropogenic eutrophication (Howarth et al., 2011). All these drivers can affect species directly or indirectly, and reduce biodiversity in an ecosystem. In addition, anthropogenic drivers, like habitat conversion and occupation as well as environmental pollution, can force a loss of biodiversity.
Species can, due to environmental changes or by the opening of pathways, enter new biogeographical regions and establish an invasive population there. In an ecosystem with high biodiversity, the resistance to invasion is much higher (Kimbro et al., 2013). In an ecosystem with many different species, resources are efficiently used and it is hard for an invading species to compete with already established allocation of resources (Hodapp et al., 2019), in other words, filled niches. This is reinforced by rare species occupying small niches, which are, therefore, no longer available for invading species (Lyons and Schwartz, 2001). From anthropogenic activities, opportunistic species can get introduced into a foreign ecosystem, including new diseases and pathogens (Crowl et al., 2008). The loss of biodiversity can open previously filled niches and alter food web structure, which facilitates the invasion of species (Lyons et al., 2005). All this can accelerate species turnover rates. Ecosystems with high species turnover rates are likely to have a reduced resource use efficiency (RUE) (Ptacnik et al., 2008). This and the new competitions from invasive species can make it hard for species to establish a stable population and optimize their RUE. This can result in a lower total biomass production (Ptacnik et al., 2008; Hodapp et al., 2019).
Not only can the presence or absence of certain species affect ecosystem dynamics, but this decline also reduces genetic diversity, which can lead to a reduced resilience or plasticity to changing environmental conditions. Therefore, species and populations would have a lower speciation potential (Brennan et al., 2019). Fewer individuals also increase the chances of inbreeding, which often negatively impacts populations (Charlesworth and Charlesworth, 1987; Charlesworth and Willis, 2009). The loss of genetic diversity can negatively impact ecological resilience during extreme events (Reusch et al., 2005). This general description across habitats may not apply to all ecosystems. Please note that the arrows shown in Figure 2 are not the only possible connections in ecosystem functioning.
Fisheries Management
Oceans ecosystem services have been estimated to represent more than 60% of the global economic volume (Costanza et al., 1997; Martínez et al., 2007). Obtaining these socioeconomic benefits calls for ecosystem-based fisheries management. This strategy focuses on the health of the marine ecosystem as well as the fisheries (Pikitch et al., 2004). Monitoring changes in marine ecosystem dynamics can provide insights into the alteration of ecosystem functions. Therefore, a cost-effective monitoring method is needed. The analysis of eDNA can help achieve this (e.g., Jerde, 2019).
Traditional and Novel Monitoring
The past decades have given us a lot of insight into fisheries and the marine realm through the use of traditional sampling methodologies. Nonetheless, like every method, traditional methods have their limitations. For example, visual methods, like UVC (underwater visual census), BRUVS (baited remote underwater video station), diving, or snorkeling are very expensive due to high personnel costs. Additionally, diving methodology is restricted to accessible habitats with clearer waters. Other methods, like bottom trawls, fyke nets, multi-mesh gill nets, or other catching methods are invasive and can be destructive to habitat structures and species. For example, bottom trawling can destroy corals and effects are often long-lasting (e.g., Althaus et al., 2009). Therefore, invasive methods may not be the best choice for use in protected sites such as marine protected areas (MPAs). Behavioral aspects, like avoidance, can as well influence the precision of monitoring techniques (e.g., Boussarie et al., 2018). Environmental DNA methods further extend this range of traditional tools to explore and monitor fisheries.
Several studies investigated the effectiveness of eDNA-based methods compared to these established methods. For example, Thomsen et al. (2012a) compared eDNA metabarcoding with nine different traditional methods (angling, day- and night-snorkeling, beach seine, fyke nets, push nets, fish pots, multi-mesh gill net, bottom trawl) to observe a fish community. They were able to detect higher or equal fish diversity with eDNA analysis than with traditional methods. Several other studies also addressed how well eDNA reflects teleost diversity compared to traditional methods (e.g., Schmelzle and Kinziger, 2016; Valentini et al., 2016; Sigsgaard et al., 2017). Boussarie et al. (2018) were able to detect more shark species with the analysis of eDNA than with UVC and BRUVS. More broadly, Stat et al. (2017) were able to detect 287 families across the eukaryotic kingdom, showing the breadth of taxa detectable by eDNA analysis. Despite the benefits of effectiveness of eDNA-based methods, monitoring with traditional methods is worthwhile because allows long-term monitoring due to comparability of datasets and provide additional information, like age and sex of the individuals, that eDNA-based methods cannot provide. Moreover, the single methods shine a light on different aspects of a biological community (Figure 3).
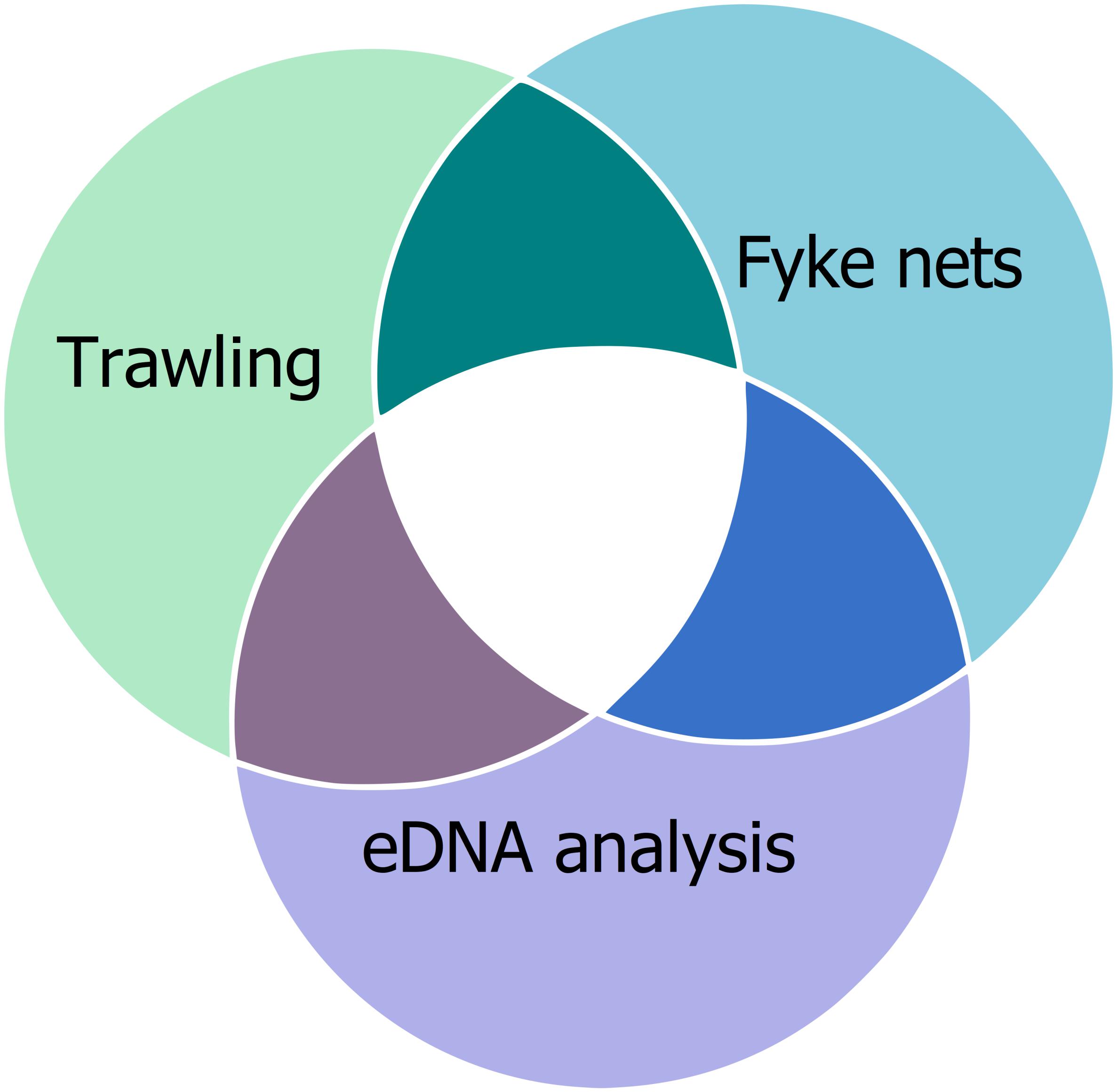
Figure 3. Toward a more complete understanding of community composition. While results may overlap between monitoring techniques, no single method entirely captures species richness. We suggest combining multiple tools to obtain a more complete picture of biological diversity, as each method has its own strengths and weaknesses. For this example, trawling, fyke nets, and eDNA analysis would be used to capture biological diversity, however, these are not the only techniques available. Methodology should depend in which questions and taxonomic groups are of interest.
The involvement of fisheries records is highly beneficial for diversity monitoring and for the monitoring of key species. Conversely, ecological theories can help to improve fisheries management as well. This win-win situation has recently improved, as fishing activities were used to test ecological theories (Jensen et al., 2011). Evaluating fisheries data can give insights in influence of density depended spatial distribution of populations (reviewed in Jensen et al., 2011). For example, Merder et al. (2020) tested the density-dependent suppression of population biomass by using data from fish traps. This study verified the effectiveness of ‘catch-and-wait’ fisheries, where caught organisms were size-selected and small organisms were placed back into the environment. But fisheries data can also help to understand ecosystem dynamics and processes, like regime shifts (ecosystems abruptly shifting into another discrete state) or changes in trophic cascades (reviewed in Jensen et al., 2011). Worm et al. (2006) compared catch data and ecological survey data and found an increased stability, i.e., higher recovery rates and lower collapsed fish/invertebrate stocks, in highly diverse ecosystems. These studies show that it is highly beneficial to use fisheries data as large ecosystem-scale experiments.
Stock Assessments
Nearly all traditional monitoring methods require significant sampling effort and personnel with high taxonomic expertise. Due to the continuous decline of taxonomic experts (Hopkins and Freckleton, 2002; Fischer, 2013), the probability of biases through misidentification increases. Taxonomic expertise is often highly specialized to a certain family or even genus due to its complexity. This specialized expertise may bias community richness results, skewing toward a particular genus, order, or family (Goodwin et al., 2017). When monitoring entire communities, identification via morphology requires high levels of expertise and time, which is often not feasible. Metabarcoding methods rely on complete and accurate libraries for environmental genetic assessment, so gaps in databases such as NCBI pose a challenge. However, as databases continue to improve, so will the detection of biological communities with eDNA. Here, we highlight some potential uses of eDNA should the databases be available for the species of interest.
A promising tool to assess fish stock sizes is the method of close kin-mark recapture (CKMR) (Bravington et al., 2016). With this pseudo-likelihood approach, it is possible to estimate stock size through fish kinship in a specific area or fishing ground. The more related caught individuals are, the smaller the stock is. CKMR is a recently developed method, combining evolutionary genetics with advanced mathematics (Bravington et al., 2016).
Fish stock assessments via catch per unit effort (CPUE) or occupancy modeling have been established for several years now and are widely used in marine management. Several studies investigated the potential of biomass/abundance estimation with correlation to the concentration of target eDNA fragments (e.g., Thomsen et al., 2016; Salter et al., 2019). Yates et al. (2019) recently conducted a meta-analysis with data from 19 studies and showed a weak correlation between eDNA concentration and abundance. In certain cases, it is possible to correlate the concentration of target eDNA fragments with the biomass of the respective target species (e.g., Lacoursiere-Roussel et al., 2016; Thomsen et al., 2016), making the implementation of CPUE in eDNA-based studies promising. Nonetheless further research is needed before eDNA concentrations can be reliably correlated to biomasses, since eDNA shedding and decay rates are highly variable between species and environmental conditions (e.g., Collins et al., 2018).
Models, such as occupancy models, try to translate scientific data into management-relevant estimations. These models estimate species occurrence and occupancy while accounting for possible non-detection or species misidentification (MacKenzie and Bailey, 2004). Site occupancy models can factor in non-detections, i.e., the patchy distribution of species. This makes site occupancy models a robust statistical framework for eDNA-based presence/absence studies (Schmidt et al., 2013). For example, Schmelzle and Kinziger (2016) compared eDNA-based detection of tidewater goby (Eucyclogobius newberryi) with a traditional method (multiple, paired seine hauls) by using multimethod occupancy modeling. They found that the detection probability using eDNA was nearly twice as high as for seining. They also found that eDNA concentration was positively correlated with CPUE (based on seine hauls) of tidewater goby (Schmelzle and Kinziger, 2016). This study shows, that eDNA analyses results can be applied in a wide range of established stock assessment frameworks.
Invasive Species
Marine management approaches also have to consider increased species invasions and their effects on ecosystem dynamics. The profound impact of biodiversity and ecosystem functions of invasive species has attracted the attention of management for over two decades (Pejchar and Mooney, 2009). The immigration of opportunistic species into a new habitat itself is a natural evolutionary process, a natural range expansion. But, due to human activities in marine ecosystems, this process has dramatically accelerated (Occhipinti-Ambrogi and Galil, 2010). The introduction of invasive species does not only have direct effects on community composition (see Section “Ecological Theory – An Overview”), e.g., through displacement of native species, but also on food web structures or even alter fundamental processes (ecosystem engineering species), such as nutrient cycling and sedimentation (reviewed in Molnar et al., 2008).
Ecosystem services can, as well, be impacted by the occurrence of non-native species, but in both, positive and negative ways (Katsanevakis et al., 2014). For example, the blue mussel, Mytilus edulis, is commercially important in human food provision. Due to the invasion of the Pacific oyster, Crassostrea gigas, into the southern North Sea, the biomass of blue mussels in this area decreased and the oysters changed the habitat structure (Kochmann et al., 2008), which had a negative effect on ecosystem services. However, the Pacific oyster is a commercially important species. This had positive effects on ecosystem services, because it offers a new economic sector. After some years, the ecosystem seemed to balance out and the blue mussel grew underneath the oyster-beds (Reise et al., 2017), benefiting from the oysters’ protection against the main predator, Carcinus maenas, and biodiversity increased (Markert et al., 2009). Thus, an introduced species has the potential to increase diversity and total biomass production of the ecosystem (Figure 2), like in the example of the oyster-beds, diversity as well as total biomass production increased (Markert et al., 2009). The invasion of species can force more efficient resource use, and, therefore, a higher ecosystem productivity, but only if the species’ traits fill a niche in the existing system (Hodapp et al., 2019). Nonetheless, invasive species more often have a negative impact on ecosystem functioning (Katsanevakis et al., 2014). If an introduced species competes with one or more native species in an ecosystem, it can decrease diversity and accelerate species turnover rates, which can lead to a decreased total biomass production (Figure 2).
The awareness of invasive species affecting ecosystems increases the need for fast, cost-effective, and standardized methods (Darling and Blum, 2007). DNA-based methods to identify species presence, within the context of species invasions, especially in their early life stage, has proven to be highly suitable (e.g., Radulovici et al., 2009). In addition, the decline of taxonomic experts can narrow the scope to certain taxa (Taylor and Harris, 2012), and, thus, might lead to a misinterpretation of invaders and their impact on ecosystem dynamics.
Using molecular methods, the presence of species can be detected solely through their DNA traces in the environment. With the analysis of eDNA, it is possible to track the spatial and temporal occurrence of species (e.g., O’Donnell et al., 2017). The incredible sensitivity of this method allows the detection of only a few individuals entering a new area (e.g., Ardura and Zaiko, 2018). For example, Marshall and Stepien (2019) were able to detect the invasive Eurasian zebra and quagga mussels (Dreissena polymorpha and D. rostriformis) across all life stages. As well, eDNA results delineated species compositions, and even population-level diversity (see Section “eDNA and Evolution”) differences among ecosystems (Marshall and Stepien, 2019). Studies investigating the spatial distribution of aquatic animals has been done for a variety of taxa (e.g., red fin perch; Bylemans et al., 2016). Defining spatial resolution of species distribution in marine systems are, to our knowledge, not proven, due to the lack of knowledge about water body movements and the degradation characteristics influencing the spread of eDNA (see Sections “eDNA and Water Chemistry” and “eDNA and Hydrodynamics”).
Pathways of Introduction
Since the removal of an already established invasive species is usually unsuccessful (Thresher and Kuris, 2004), monitoring and regulation of introduction pathways can prevent species invasion (Molnar et al., 2008). Opportunistic species can enter new habitats in a variety of ways. Some of the major pathways are unbarred via globalization and increased transport across the world, for example, larval transport via ballast water or organismal transport through hull fouling (Molnar et al., 2008). One of the challenges of ballast water monitoring is that many of these invasive species may be in larval form, which makes them difficult to identify morphologically (McManus and Katz, 2009). This is especially important to consider in light of declining taxonomic expertise (Hopkins and Freckleton, 2002). Species identification via metabarcoding with well-curated databases can be implemented by monitoring ballast waters with eDNA to improve the effectiveness of invasion prevention.
Some of the first studies to metabarcode eDNA/bulk samples from ballast water used universal CO1 and RuBisCo markers from a ship traveling from Germany to South Africa (Ardura et al., 2015; Zaiko et al., 2015a,b). Multiple taxa were identified to assess the biological communities traveling in ballast water ecosystems, including algal and mollusk species which can pose an invasion risk (Ardura et al., 2015; Zaiko et al., 2015a). Other studies have also used eDNA methods to detect Quagga (Dreissena bugensis) and Zebra mussel (D. polymorpha), but found that light transmission spectroscopy was more effective for detection (Egan et al., 2015). While eDNA methods alone may not be ready to become the only test for industrial-scale usage, these genetic methods of presence identification are likely to be a good complement to current methods (Zaiko et al., 2015b). This may especially be true if eDNA metabarcoding proves to be economical, fast, and can be standardized across labs.
Anthropogenic habitat engineering can also open the floodgates for species invasion. One of the most famous examples was the construction of the Suez channel that connects the Red Sea and the Eastern Mediterranean Sea in 1896. With this direct connection between the Indian Ocean and the Mediterranean Sea, more than 300 species migrated to new locations (Por, 1971; Bentur et al., 2008). This rapid breakdown of distribution barriers and the resulting acceleration of species turnover issued a challenge for ecosystem stability (Figure 2).
Another opportunity for species invasions is via aquaculture and fish farms, especially when species are cultured in an area in which they are not native. A recent example is the invasion of the Pacific oyster (Crassostrea gigas) into the North Sea. Due to increased winter temperatures, induced by climate change, escaped oyster larvae from the farming grounds established invasive populations (Diederich et al., 2004). This example also shows that anthropogenically driven species invasion has more than one facet that needs to be considered (here, climate change and aquaculture). But not only aquaculture needs scrutiny; nearly all occasions where living animals are transported comprise a high potential to open pathways for invasive species, like the trade of live seafood or the introduction of exotic pets (e.g., the aquarium trade) (Molnar et al., 2008).
How species enter new habitats is one of the most crucial areas for marine management. Geographically, pathways of invasion can be reconstructed via population genetics (Estoup and Guillemaud, 2010). For example, Hänfling et al. (2002) found that the Chinese mitten crab (Eriocheir sinensis) population, invaded in San Francisco, CA, United States, was founded by a single invasion event from European, not Chinese, populations by analyzing population genetic data. Increasingly, evidence has built up around eDNA population genetic studies (see Section “eDNA and Evolution”) for improving our understanding of invasion pathways.
Marine Conservation
The conservation of biodiversity in highly endangered marine habitats is crucial due to high extinction rates (IPBES, 2019). In marine management approaches, stakeholders try to regulate the occurrence of certain species (e.g., invasive species), or the size and structure of stocks. Marine conservation approaches, by contrast, rely on the paucity of human influence, allowing nature to restore itself without much active management. A detailed knowledge about the spatial and temporal distribution of marine species is necessary for successful conservation.
Marine Protected Areas
Areas which are protected from most anthropogenic impacts, allow ecosystems to recover and stabilize. Modern protected areas were set up during the last 200 years in all habitats, but there is evidence that the ideas for protected areas are roughly 2000 years old (Phillips, 2004). In the 1970s, the establishment of protected areas increased exponentially, trying to restore biodiversity and maintain ecosystem services, especially in marine systems (marine protected areas, MPAs). Positive effects of MPAs on socioeconomic value has been widely reported (e.g., McClanahan et al., 2006).
Monitoring biodiversity in MPAs and the surrounding area is one of the cornerstones used to evaluate the success of protection effort. So far, the abundance of some key species was estimated with traditional, mostly invasive methods, such as trawling. These invasive methods can impact rare or endangered species. The use of eDNA to evaluate diversity, or the occurrence of single species, can improve non-destructive monitoring in MPAs. But traditional, mostly invasive methods provide information that eDNA analysis cannot. For instance, size and gender of single individuals allows researchers to estimate the structure and health of a stock. Nonetheless, eDNA metabarcoding reveals great benefits, like the identification of rare species with lower sampling effort, potential abundance estimations, and species composition.
Rare and Endangered Species
The occurrence of rare species is one of the major criteria for the establishment of marine protected areas (MPAs). Another one is high diversity, so called diversity hotspots. Most of the time, rare species occur less often in areas where diversity is high (Prendergast et al., 1993). Naeem (1998) already proposed that rare species should have a large impact stabilizing ecosystem functions, based on ecological and engineering theories. Recent studies supporting this theory, like Mouillot et al. (2013), found that 98% of locally and regionally rare fish species highly support ecosystem function in coral reef systems. The maintenance of ecosystem functioning not only relies on high diversity, but on species-specific contributions to certain functions. Studies showed, for example, that the removal of rare species decreases the invasion resistance of an ecosystem (Figure 2; Lyons and Schwartz, 2001). The more rare species go extinct, the higher acceleration of species turnover will be.
The incredible sensitivity of eDNA analyses are not only beneficial to detect invasive species at an early stage, but also rare and endangered species that can easily and non-invasively be monitored. For example, Parsons et al. (2018) was able to characterize the stock structure of the elusive harbor porpoise (Phocoena phocoena) from inland waters of southeast Alaska. Beyond species presence, eDNA can identify the effects of invasive species on the distribution of native species (Wilcox et al., 2018), or even the distribution of invasive species itself and the efficiency of management efforts (Robinson et al., 2019).
Some studies indicate that eDNA-read abundance correlates either with the biomass or with the number of individuals (e.g., Salter et al., 2019). One of the first studies in the marine environment to correlate eDNA-read abundances with fish abundance, caught by trawling, was conducted in Greenland waters (Thomsen et al., 2016). However, the results were more nuanced. Some species had a more significant correlation between read abundance and trawl abundance than other species. This could be because many factors influence detectability and read abundance in an eDNA sample. This includes different shedding rates, according to life cycle stage, or different persistence rates, due to environmental conditions. Additionally, low density species may be detected stochastically depending on sampling effort, as is the case with all detection methods. How samples are obtained and preserved can interfere with certain lab procedure steps, thus, affecting detectability and read abundances (Mauvisseau et al., 2019). Different lab procedures, like extraction protocols or preservation, but also primer biases can cause variation in read abundances (e.g., Kelly et al., 2019). Biologically, eDNA shedding and persistence rates are highly species and life cycle specific (Sassoubre et al., 2016; Ahern et al., 2018). For example, higher biomass caused higher shedding rates of eDNA from Japanese jack mackerels (Trachurus japonicus) in a tank experiment (Jo et al., 2019). Thus, part of the difficulty in relating abundance to eDNA is that species-specific relationships will need to be teased apart.
Such discrepancies require a lot of preliminary work to evaluate accuracy when using a single species approach. Another possibility is the relative quantification of a species within the community (e.g., Valentini et al., 2016). The emerging growing technologies in the field of high throughput sequencing allows us to describe whole communities in one sequencing run (see Section “Pathways of Introduction”).
However, eDNA is not currently used for providing concrete estimates of abundance, such as biomass or individual count (Hansen et al., 2018; Yates et al., 2019). To link eDNA to abundance in a marine environment, more studies need to be done to accurately model how eDNA moves through the water column. While this has been done for freshwater environments, the marine system presents more complex challenges, for example, tides and currents must be taken into account (Kelly et al., 2018; Lacoursiere-Roussel et al., 2018; Andruszkiewicz et al., 2019; Levi et al., 2019). In addition, eDNA cannot yet provide information like fish size and age, or gage hormone levels (Hansen et al., 2018).
Over time and increased research, eDNA may be able to provide information on species shifts and changes in species presence. With further developments, eDNA may become a tool useful for monitoring, more than simply detecting the presence of important species. Although fisheries research can start to harness eDNA to monitor species presence now (Thomsen et al., 2016), challenges relating to abundance and life-history descriptions need to be overcome to fully use eDNA for fisheries management.
Lifecycle Aspects
Over the past few decades, the implementation of animal behavior research into conservation considerations has improved the understanding of anthropogenic impacts on biodiversity. An organism’s adaption to stressors, including anthropogenic stressors, is based on evolutionary processes, altering species behavior which might even affecting community composition (reviewed in Berger-Tal et al., 2011). The habitat alteration of spawning grounds, for example, can cause a spatial shift or a decline of species abundance; both can lead to local community shifts across trophic levels. Gosset et al. (2006), found that population size was significantly negatively affected by anthropogenic actions, due to the alteration of migration behavior. Annual or seasonal movement of marine species is often triggered by at least one of the following reasons: mating, food availability, or climatic aspects (Lascelles et al., 2014). Additionally, life stages can force migration behavior. Juvenile or larval stage of marine species are often part of the zooplankton community. In this case, migration often takes place on a daily cycle. Berry et al. (2019) was able to identify 245 families of eukaryotic zooplankton with eDNA metabarcoding and the majority of them were at a larval stage. Another example is the vertical migration of Antarctic krill (Euphausia superba) known for nocturnal migration to feed on plankton at surface waters and diurnal migration to deeper waters for predator avoidance (Croxall et al., 1985). Relating to this vertical migration of krill, fish stocks follow this migration pattern to feed on (Kaartvedt et al., 1996). Life cycle migration in the Pacific Salmon (Oncorhynchus spp.) was detected with an eDNA analysis in a time series in AK, United States (Levi et al., 2019). Recently, conservation approaches tried to include such spatial variation due to life cycle stages, to improve the protection of endangered species (see Section “Implications for Marine Conservation”). Recognition of food web and life cycle migration aspects is a critical point to ensure successful conservation and eDNA analysis also be applied here.
Implications for Marine Conservation
Twenty-one percent of marine migrating species are considered to be threatened (Lascelles et al., 2014). Therefore, efficient monitoring of species diversity and richness in MPAs are highly useful to evaluate the success of protection. eDNA-based analysis can help to improve monitoring in this area, especially for rare and endangered species. However, migration patterns needed to be considered. Behavior-based modeling of spatial distribution and habitat use improves conservation success and should be one of the key tools for marine conservation (Norris, 2004). In recent years, the idea of pop-up protected areas has emerged (Runge et al., 2015). Knowledge about migration routes and times allow for protecting certain areas at that time, such as when endangered species migrate through (Runge et al., 2015). This terrestrial approach is highly promising for conservation and meets economic needs. We suggest this concept be used in marine conservation as well (Figure 4).
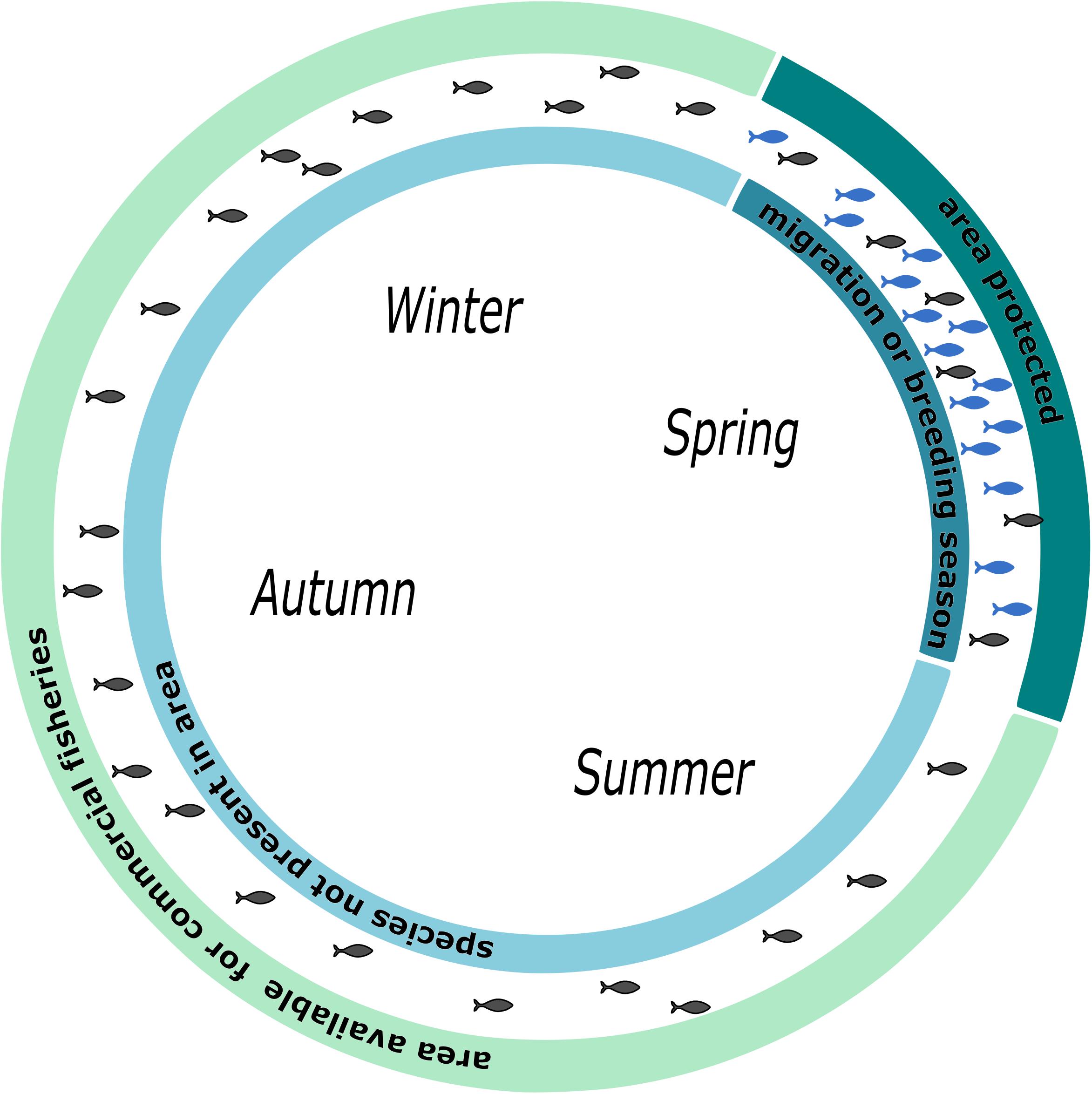
Figure 4. Schematic illustration of pop-up protection areas in marine realm. Differential usage of a seascape by fisheries and conservation efforts. During the breeding season, the area is protected from fishing. Other times of the year, the area is open for fishing. Alternatively, a seascape could be protected during a migratory season.
Anthropogenic Influences on Marine Communities
More broadly than fisheries decline, there is a need to monitor changes in the biodiversity of communities as they impact ecosystem services (Hooper et al., 2005) (see Section “Marine Management and Ecosystem Functioning,” Figure 2). Community composition can be described with a variety of indices (Simpson, 1949; Pielou, 1966; Hill, 1973; Lande, 1996), traits (Litchman and Klausmeier, 2008; Weithoff and Beisner, 2019), and phylogenetics (Mouquet et al., 2012; Tucker et al., 2017) to predict how changes in community assembly over space and time may affect ecosystem function. To inform these models, individuals were captured through a variety of traditional methods such as nets (Siegenthaler et al., 2019), video (Stat et al., 2019), transects (Uthicke et al., 2018), and quadrats (Currier et al., 2018). However, because of the aforementioned decline in taxonomic expertise and effort of collecting samples (Hopkins and Freckleton, 2002), eDNA analyses, especially eDNA metabarcoding, are emerging as a genetic method for data acquisition to complement traditional sampling and can foster large scale community monitoring.
Aquaculture and Seascape Shifts
Seafood production through aquaculture and fish farms more than doubled since the 1980s (Asche and Smith, 2018). At least since the 1980s, fish farm impacts on the surrounding ecosystem have been well documented (Brown et al., 1987). These biological changes can now be quantified using eDNA-based methods as well as traditional methods (e.g., Stoeck et al., 2018a). One instance examined foraminiferal biodiversity using protist eDNA metabarcoding primers in areas both close and far from salmon farms in Scotland (Pawlowski et al., 2014). Samples farther from salmon nets showed greater species richness of foraminiferal communities compared to samples closer to salmon farms (Pawlowski et al., 2014). Other Scottish and New Zealand biodiversity investigations found similar results with bulk sampling for bacteria and protists (Dowle et al., 2015; Pochon et al., 2015; Stoeck et al., 2018a,b). New Zealand’s Chinook salmon farms (Oncorhynchus tshawytscha) also showed that high waterflow sites had higher foraminiferal diversity than low waterflow sites, perhaps indicating higher water flow is better for the fish farm ecosystem (Dowle et al., 2015; Pochon et al., 2015). In British Columbia, foraminiferal alpha diversity was lower close to salmon cages compared to more distant reference sites, though beta diversity showed impacts may differ between farms (He et al., 2019). Importantly, shifts in foraminiferal communities were investigated with both eDNA and eRNA, the latter providing a more recent snapshot of the community at large (Dowle et al., 2015; Pochon et al., 2015). The use of eRNA provides a better proxy for modern communities due to eRNA’s shorter half-life in an environmental setting.
These impacts on biodiversity and community composition at the base of marine food webs are likely to be transferred through all trophic levels (Figure 2). Environmental DNA may persist for a longer period of time when using sediment as the environmental substrate (Turner et al., 2015), giving the opportunity to track community shifts over time (Armbrecht et al., 2019). Occurrence of phenological mismatches are one major driver accelerating species turnover rates (Figure 2). This, in turn, may decrease biodiversity as well as ecosystem biomass production.
Environmental Pollution Events
Not only salmon farms may shift surrounding community composition. Oil and gas operations also showed differing benthic community compositions when compared with reference sites (Laroche et al., 2018; Cordier et al., 2019). While all these interactions have been documented with other methods (Kalantzi and Karakassis, 2006), eDNA is an effective environmental impact monitoring tool for community biodiversity, especially for harder to sample meiofauna (Olsgard et al., 1998; Lejzerowicz et al., 2015; Stoeck et al., 2018b). eDNA has also been proposed to monitor mining areas, especially deep-sea mining, although this has yet to be extensively researched (Fernandes et al., 2018; Billett et al., 2019). More research is needed to apply this framework to bigger, free-swimming macrofaunal assessments. Their genetic signatures may not be as strong given the larger home ranges of many macrofaunal marine taxa (Boerder et al., 2019).
Environmental DNA can also be applied to monitor the aftermath of unexpected events, such as oil spills. After such events, eDNA can aid in quantifying the effectiveness of clean-up efforts to characterize ecosystem resilience and health (Ellis et al., 2012). For example, after the Hebei Spirit oil spill, eDNA monitoring was used to determine how well the micro- and meso-organismal communities bounced back (Xie et al., 2018). Immediately following the oil spill, bacteria which degraded hydrocarbon and algal families were stimulated by oily sediments, while protist communities changed (Xie et al., 2018). After five years, metazoan communities with less contaminated sediments started shifting toward diversity profiles similar to reference sites (Xie et al., 2018). Another example showed a shift in communities before and after the Deepwater Horizon oil spill (Bik et al., 2012). Before the spill, a high diversity of metazoans was present and DNA-based diversity was dominated by nematodes (Bik et al., 2012). In contrast, after the spill, fewer metazoan taxa were recovered from DNA reads and diversity was dominated by fungal reads (Bik et al., 2012).
By using eDNA metabarcoding to monitor meiofaunal communities, it is possible to quickly gain a sketch of how these events shift biological communities. In the future, eDNA methods may become more widely applied to a variety of these unexpected but community-shifting events, such as marine algal blooms or extreme environmental events (Dhar et al., 2015). For example, hurricanes can reduce coral cover by an average of 17% a year (Gardner et al., 2005). Marine heatwaves may also cause shifts in community composition (Berry et al., 2019). Such extreme environmental events are happening with greater frequency due to shifting climate (IPCC, 2014). Beyond the immediate monitoring of after effects, if areas of interest – commercial, recreational, conservation, or otherwise – are consistently monitored, then long-term datasets can start to be built and long-term patterns may emerge (Berry et al., 2019). From recognizing natural recovery patterns, management of these areas post-impact could potentially hasten or direct recovery.
Water Quality Assessments With eDNA
Anthropogenic activities beyond large events, such as oil spills, often lead to a change in chemical composition of aquatic environments. This starts with the discharge of nutrients leading to increased eutrophication of coastal areas (e.g., Rabalais, 2002), which in turn changes primary production and species composition (Figure 2). Graham et al. (2019) used a Bayesian network modeling approach to evaluate water quality in estuaries and describe community structure. Another approach elucidated the functional ecology of groundwater fauna, using eDNA-based community analysis as well as eDNA stomach content analysis (Sacco et al., 2019). The understanding of groundwater ecology and the shifts in community composition and functioning it can detect, is highly important for human well-being under the light of preserving water supply.
In ecotoxicological research, eDNA approaches can be implemented (Zhang, 2019). For example, the release of endocrine disrupting chemicals can facilitate phenological shifts, which may impact whole ecosystems (e.g., Segner et al., 2003) community response to toxic substances can be evaluated. Biodiversity changes caused by pollutants may affect ecosystem dynamics (Zhang, 2019). Single-species abundance changes may also be targeted with qPCR and specific probes. Thus, monitoring with eDNA-based techniques, whether it be metabarcoding for community or targeting a single species, can aid in discerning patterns of community-level shifts in ecotoxicological research and can inform management efforts.
Interdisciplinary Future of eDNA Monitoring
Currently, eDNA analysis can confidently detect species presence and describe species composition. The use of eDNA-based methods has many advantages, but the methods still require more research to overcome challenges and expand the scope of its use (Goodwin et al., 2017; Cristescu, 2019). We consider harnessing the wide potential of eDNA as a tool to answer evolutionary and ecological questions beyond presence and abundance by linking eDNA with other existing areas of research (Figure 5). One area would be to fully develop eDNA for discerning evolutionary change, including for population genetic and epigenetic markers (Zhao et al., 2018; Adams et al., 2019; Sigsgaard et al., 2020). Other avenues may include adding eDNA to suites of ecosystem monitoring tools, such as combining eDNA with water chemistry to obtain a more complete idea of how abiotic and biotic factors are connected within an ecosystem over time and space (He et al., 2019; Zhang, 2019). Already, eRNA has been used in conjunction with eDNA to snapshot biodiversity at shorter time scales, and potentially to understand the biological processes present (Pochon et al., 2017; Zaiko et al., 2018; Cristescu, 2019). Lastly, we briefly touch upon the potential of combining hydrodynamics with eDNA methods and the increased interest of monitoring eDNA autonomously or by remote sensing methods (Scholin, 2010). Automation may aid in consistency of monitoring and reproducibility of research (Dunn et al., 1993; Pomati et al., 2011). While fine-tuning eDNA remains for ecological questions, eDNA shows flexibility and promise as a tool for ecosystem monitoring when coupled with other methods.
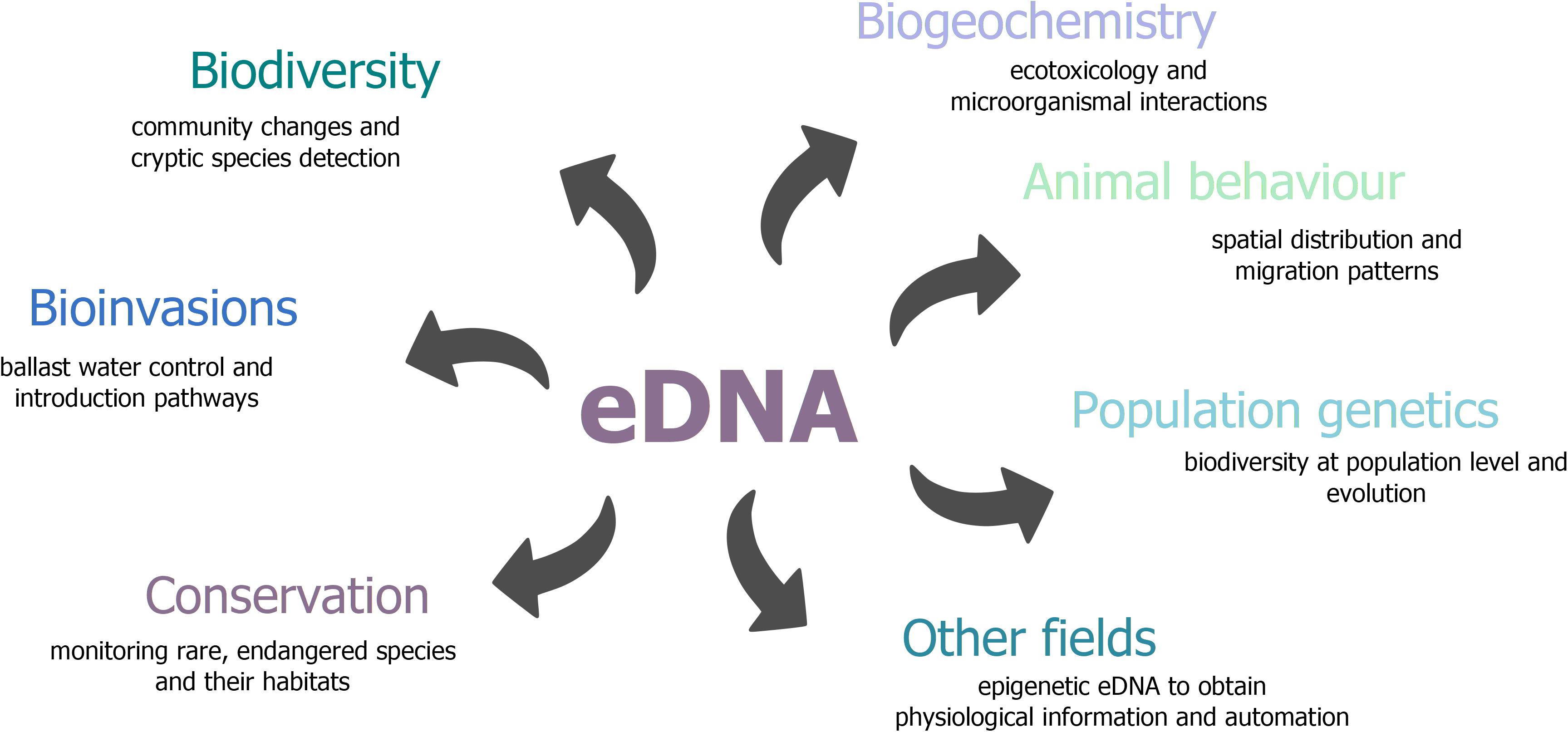
Figure 5. Overview of research fields that can benefit from eDNA-based approaches. Outlined are a few potential applications.
eDNA and Evolution
Population-Level Variation With eDNA
Modern eDNA analysis is increasingly being explored for genetic monitoring. Currently, groundwork is being laid for using eDNA to describe population genetic variation, mostly in marine environments (Adams et al., 2019). Recent work has focused on the ability to quantify haplotype ratios for quagga and zebra mussels (Marshall and Stepien, 2019). From aquarium and field experiments, there was a significant relationship between observed and expected haplotype ratios (Marshall and Stepien, 2019). This finding holds promise for using eDNA for both the ability to confirm existing phylogenetic relationships, discovering new haplotypic variation, and quantify this variation. However, more tests using other ecosystems and species will be needed to confirm this trend, but given the recent success, eDNA for detecting population-level variation may be on the horizon.
To be able to use eDNA for population genetic questions will require more research. First, some population genetic theory cannot be tested with eDNA if individuals cannot be identified (Adams et al., 2019). Many arguments for eDNA population genetic usage reflect that of pooled sequencing (poolseq), however, it is difficult to ensure that plentiful and equal amounts of individuals per species are in an eDNA sample (Schlötterer et al., 2014). To understand how haplotypes are distributed in a marine water column, the “ecology” of eDNA (see Section “eDNA and Water Chemistry”) needs to be investigated. With this knowledge, genetic variation based on population-level diversity could be investigated for more marine species (Barnes and Turner, 2016; Marshall and Stepien, 2019). While evidence suggests correlation between eDNA population-level genetic variation and traditionally sampled genetic variation, this remains to be tested under other conditions (Marshall and Stepien, 2019). It is likely that bioinformatic filtering parameters may need to be system-specific and fine-tuned for eDNA population genetic research, rather than a one-size-fits-all approach (Alberdi et al., 2018; Turon et al., 2019). Furthermore, as a majority of eDNA population genetic studies have focused on mtDNA, if nuclear variation cannot be obtained, eDNA population-level variation may not be as informative as traditional genetic sampling (Elbrecht et al., 2018; Parsons et al., 2018; Adams et al., 2019). If population genetic eDNA technique is further developed, this can potentially open up the monitoring of within-species genetic diversity for multiple species at the same time (Stat et al., 2017; Elbrecht et al., 2018).
Epigenetic eDNA
Advantages of epigenetic eDNA – What is it and why do it
The integration of epigenetics and eDNA has the potential to allow researchers to quantify genetic variation in eDNA samples, and, thus, make use of basic population genetic frameworks (Zhao et al., 2018; Sigsgaard et al., 2020). Epigenetics, modifications of DNA which are not alterations to the DNA sequence itself, acts as an interface between the environment and genetics because modifications are made in response to the environment. While epigenetic variation such as methylation has yet to be extracted from eDNA sources, this presents an intriguing opportunity because information not normally obtained from eDNA could be detected. Heritable epigenetic changes could be used as measures of relatedness at a population level for distinguishing and comparing populations or individuals (Fraser et al., 2012; Zhao et al., 2018) and even individuals (Fraga et al., 2005).
Additionally, epigenetics reveal aspects of populations that eDNA genetic analyses alone cannot currently discern (Fraga et al., 2005). Methylation patterns differ with age and sex, which could be of interest for tracking population dynamics. Evidence also exists for different methylation patterns based on environmental factors, such as nutrition, temperature, and toxins (Gokhman et al., 2017). Epigenetics may also play a role in disease and pathogen resistance (Feinberg, 2018). Through obtaining epigenetic information, these environmental effects on target species could be determined. Moreover, due to the role of methylation in transcription, differences in gene expression between populations could be examined (Nätt et al., 2012). Variable expression from epigenetic markers could affect how organisms are locally adapted or hint as to what traits may be caused by phenotypic plasticity (Sahu et al., 2013; Aller et al., 2018; Yang and Andrew Pospisilik, 2019). Because eDNA is a bulk sample, epigenetic information for multiple species could be obtained from the same sampling event. Over time, ecological and evolutionary patterns may emerge at the community level. However, much more research, including proof-of-concept work, would need to substantiate any of these ideas before epigenetic eDNA (epi-eDNA) can be used for monitoring.
Epigenetic eDNA (epi-eDNA) challenges and how to address them
While analyses of both epigenetics and eDNA are powerful in their own right, epi-eDNA analysis likely faces multiple challenges before the approach can be routine and practical for in-field use. One challenge would be to separate out patterns of methylation, as different tissue types contain different patterns of epigenetic variation, even within the same individual (Christensen et al., 2009). Metastable epialleles, loci that are differently methylated between individuals but have low variability within the same individual, could be targets of epi-eDNA (Rakyan et al., 2002). This would require methylation maps across different tissues and individuals, which is not currently very common for non-model species.
Beyond finding a suitable target, epigenetic eDNA techniques would likely require well-preserved epi-eDNA, which may present technical challenges. However, isolating DNA from degraded sources such as ancient DNA is possible and has been done using bisulfate sequencing (Smith and Ryckman, 2015; Gokhman et al., 2016, 2017; Zhenilo et al., 2016). Alternatives to harsh bisulfate sequencing using enzymatic deamination methods have also been developed and these may be favored for low concentration epi-eDNA samples. Once samples have been taken, separating target cells from non-target environmental debris may prove challenging and expensive, though could be possible with cell sorting techniques such as a fluorescence-activated cell sorter (FACS) for tissue-specific and species-specific cells (Birnbaum et al., 2005; Smallwood et al., 2014). However, FACS is more efficient with live cells, as fluorescent markers will need to bind on extracellular proteins which could degrade over time in the environment. There may be limitations to what inferences science can make when only obtaining a snapshot of a community based on live cells present in a sample. Once cells have been obtained, single-cell epigenomics sequencing methods are possible, though more development is needed (Guo et al., 2013; Clark et al., 2016). Untangling the sequencing output with bioinformatics for epi-eDNA, beyond initial laboratory work, may present a challenge. Sorting reads according to species, individual, and/or tissue type may prove even more difficult than standard eDNA population genetics. As yet, no bioinformatic pathways have been successfully established for epi-eDNA using either single-species or metabarcoding approaches, but this area of eDNA research is ripe with possibility.
eDNA and Water Chemistry
Ecosystems are composed of an interplay between biotic and abiotic factors. Describing how abiotic factors influence and constrain biotic communities, and how these in turn shape abiotic surroundings is just one facet of ecosystem ecology. For example, abiotic factors can shape animal behavior (Kleinhappel et al., 2019), and animal behavior changes the environment (Deegan, 1993; Roman et al., 2014). If eDNA is to be analyzed to describe these processes, then teasing out how both biotic and abiotic factors shape the shedding and degradation, the “ecology” of eDNA is important.
One factor to consider may be that organismal behavior is affected by seasonal changes. Seasonal activities likely affect the amount of eDNA shed by some organisms, for instance, due to increased movement, gametes release, or life cycle stage, which consequently influences eDNA detectability (Klymus et al., 2015; Spear et al., 2015; Bylemans et al., 2017). Indeed, the migration of some organisms shows different eDNA signatures during different seasons, even indicating variation in community assemblage (Stoeckle et al., 2017; Xu et al., 2018).
In addition to the release of eDNA, seasonally variable abiotic factors such as temperature and UV may also speed or slow degradation (Strickler et al., 2015; Buxton et al., 2017). There have been mixed results on the influences of temperature and UV on eDNA degradation and evidence suggests that perhaps the main influences on eDNA degradation may not come from these sources alone (Strickler et al., 2015; Buxton et al., 2017; Mächler et al., 2018; Salter, 2018). Notably, microorganisms are an influential part of marine communities. Besides their broad role in the trophic cascades, ecosystem services, and gut microbiota, they influence the degradation of eDNA as well. In the environment, when microorganisms are nutrient limited, they will consume eDNA for phosphorous and, thus, speed up the degradation process (Salter, 2018). Additionally, there is evidence that eDNA can travel to areas through diet of predatory animals (Guilfoyle and Schultz, 2017). Therefore, further characterizing the role microorganisms play on eDNA as well as how that interacts with abiotic factors are important pathways to elucidate.
With an increase in atmospheric CO2 concentrations, ocean acidification and global warming likely also influences eDNA shedding and degradation. The increase of dissolved carbon dioxide and subsequent reduction of bioavailable calcium carbonate (CaCO3) influence behavior of marine organisms, which may then influence eDNA release (Mostofa et al., 2016; Nagelkerken and Munday, 2016). For example, crabs in higher pCO2 levels decreased foraging behavior (Dodd et al., 2015) and low pH can influence respiration rates in scallop (Liu and He, 2012). Increasing temperature has shown to increase metabolic rates as well (e.g., Eder et al., 2018). In combination with low food availability, this effect can even be strengthened (Eder et al., 2018). Besides indirectly influencing shedding rates of eDNA, evidence exists that very low pH can increase eDNA degradation, though effects are most likely to be seen with an increase in temperature and UV (Strickler et al., 2015). Potentially, ocean acidification should be considered when using eDNA as a tool for monitoring as high variability in pH may influence shedding rate or degradation, and, therefore, species detection.
Besides these influences on eDNA, a suite of geochemical corollaries is often used to describe ecosystems health. Indeed, a large body of literature has been dedicated to analyzing these interactions, and eDNA methods are only one technique used to describe biological community diversity within a suite of tools (Levin et al., 2009; Pan and Wang, 2012; Lowe et al., 2016; Mostofa et al., 2016). For example, planktonic and microbial communities may differ depending on differing dissolved organic matter found in an environment (Kujawinski, 2011; Osterholz et al., 2016; Laroche et al., 2018). A healthier, more diverse community correlates with increased dissolved oxygen (He et al., 2019). Flow of water may also influence dissolved oxygen content and, thus, community composition (Pochon et al., 2015). Besides dissolved oxygen, patterns of biodiversity have also been examined when they correlate with trace metals, for instance, toxic sulfides (He et al., 2019). Indeed, less alpha diversity was found with higher sulfide concentration using eDNA methods (He et al., 2019). Under harsher conditions, more sensitive fauna may not be as supported, and, thus, ecosystem function overall may decline (Lohrer et al., 2012). Through better understanding the interactions between biotic communities and these geochemical factors, we can better assign the ecological function of these communities and ecosystems.
eDNA and Hydrodynamics
Hydrodynamic modeling allows for the characterization of oceanic currents and, therefore, can help to answer ecological questions. For example, passive drift of larvae or other planktonic organisms can be revealed (Skliris and Djenidi, 2006; Leis, 2007; North et al., 2008). Combining such insights with genetic data, findings can resolve complex issues of ecological research. The genetic variation in the kelp Laminaria digitata along the Irish coast were underpinned by hydrodynamic distances (Brennan et al., 2014). Populations that were connected through currents had higher genetic similarity than populations that were separated by currents, even though sometimes the non-connected populations were located closer together. Even the occurrence of hybrid zonation, for example, hybrids between the mussels Mytilus edulis and Mytilus trossulus in the Baltic Sea, can be revealed by combining genetical approaches with hydrodynamic modeling (Stuckas et al., 2017).
There has been some work done to model eDNA transportation in freshwater riverine networks and in marine settings (Carraro et al., 2018; Andruszkiewicz et al., 2019; Fremier et al., 2019). Environmental DNA can be used to track organismal movement such as migration, as well as spatial patterns (Levi et al., 2019). Nearshore tides may only marginally influence the eDNA signal (Kelly et al., 2018; Murakami et al., 2019), most likely, the eDNA signal stays within 30 m of an organism in a marine setting within a few hundred meter from the shoreline (Murakami et al., 2019). Andruszkiewicz et al. (2019) modeled eDNA transportation in a distance of 15-20 km from the shoreline in Monterey Bay, CA, United States. They could show that eDNA is likely to be transported up to 40 km from the source. Various studies showed that decay rates are higher close to the shoreline than offshore, but variation in half-life times are quite large in the different study systems (Collins et al., 2018). Due to global variations of hydrodynamic patterns, further combined modeling of hydrodynamics and eDNA persistence is needed. This can be used to better link eDNA and animal activity.
Animal behavior itself can also alter the occurrence/persistence of eDNA. Currents and tides influence the activities of organisms, such as upwellings of nutrient-rich waters (Ryther, 1969; Block et al., 2011). For example, seasonal and vertical migration may be influenced by hydrodynamic movements which influence animal behavior (Batchelder et al., 2002; Croll et al., 2005). These behavioral changes may, in turn, influence eDNA signal. However, one possible false positive source that has not been quantified is the influence of dead organism eDNA in a marine setting (Kamoroff and Goldberg, 2018). Indeed, eRNA has been suggested for examining a shorter time frame (Pochon et al., 2017), though more recent work has suggested that aqueous eDNA stays in the marine environment on the scale of hours (Baker et al., 2018; Murakami et al., 2019). These studies highlight the need to be conscious of how biotic factors shape eDNA once released into the environment.
eDNA and Automated Sampling
Recent developments in technology have led up to an interest in automated eDNA sampling. Already, sampling has been assisted with automated, robotic technology to detect the presence and density of target organisms (Marques et al., 2013; Stern et al., 2015). Unmanned aerial vehicles can already collect whale breath samples for respiratory tract microbiome analyses (Pirotta et al., 2017) and collect water samples (Ore et al., 2015). One advantage of using robotic technology is the ability to leave an automated sampler out in the field for a period of time while still reliably taking samples at regular intervals, standardizing data collection (Marques et al., 2013). This reduces spatial and temporal variation in the data, which, for difficult-to-sample areas such as the open or arctic oceans, may be beneficial. Sampling like this has already been done for microbiological samples (Scholin, 2010). These systems can be applied for sampling eDNA. Yamahara et al. (2019) sent an autonomous underwater vehicle with an environmental sample processor out in Monterey Bay. In addition, they performed traditional eDNA samples and compared the results, without significant differences. This first approach needs to be tested and quantified across multiple ecosystem dynamics for reliable results.
Besides presence-absence data, more measurements could be obtained using unmanned vehicles. For example, automated underwater qPCR (quantitative PCR, quantifies the abundance of target DNA fragments) sampling has been used to detect cyanobacteria abundance (Preston et al., 2011; Ussler et al., 2013). Based on this development, automated probe-based qPCR for eDNA could be developed for identifying specific species, such as endangered or invasive species of interest. Further research would be needed to create and sequence libraries autonomously in a field setting. The advent of MinION nanopore sequencing may aid in this development given their already successful sequencing results under remote field conditions (Pomerantz et al., 2018; Truelove et al., 2019). Once set up, the maintenance of such a system may reduce personnel hours spent in the field or in difficult-to-sample places, such as the open oceans or hydrothermal vents.
Another elegant solution is to use natural sampling units. Mariani et al. (2019) successfully used the filtration characteristics of sponges to capture eDNA from biotic communities of the surrounding area. Using natural structures may be a more sustainable research approach as opposed to high-tech instruments, which are likely costly and contain plastic that degrades and pollutes. Using these natural solutions may help to protect our oceans from further pollution such as microplastics.
Conclusion
Understanding changes in biodiversity and its impact on ecosystem services is just beginning. More data are needed to fully understand complex ecosystem processes (IPBES, 2019). The use of eDNA-based methodologies can offer a wide range of data collection opportunities, from monitoring species diversity, discerning diversity not captured by traditional methods and observing changes in community composition. The addition of eDNA data into spatio-temporal ecological modeling frameworks can help elucidate species distribution and migration patterns. If coupled with diversity and ecosystem data, ecological patterns and processes can be better understood. Thus, improved predictions regarding changes in species composition and its effect on ecosystem services can be made. Furthermore, the integration of life history characteristics of species can improve conservation success. Environmental DNA-based methods have a great potential as a cost and time saving methodology. When integrated into an interdisciplinary context, eDNA offers a variety of possible applications (Figure 5) with improved efficiency in contrast to traditional methods. Currently, the field is rapidly exploring and expanding into population-level data from eDNA with implications for discerning evolutionary patterns. Future developments may include a foray into epi-eDNA to fully utilize all information obtainable from DNA molecules. This may give us a lens into the patterns and processes of genetic change within and across species. The continued development of eDNA may prove valuable for answering ecological and evolutionary questions efficiently and effectively.
Author Contributions
YS had the idea, wrote the sections “Marine Management and Ecosystem Functioning” and “Marine Conservation,” and created the figures. CIMA wrote sections “Anthropogenic Influences on Marine Communities” and “Interdisciplinary Future of eDNA Monitoring.” Sections “Introduction” and “Conclusion” were written jointly. Both authors contributed to the article and approved the submitted version.
Funding
This study was carried out in the framework of the Ph.D. research training group “The Ecology of Molecules” (EcoMol) supported by the Lower Saxony Ministry for Science and Culture, Germany. This research was also supported by the University of Otago Doctoral Scholarship awarded to CIMA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thank you to Dr. Michael Knapp and all reviewers for their comments. We especially want to thank ICYMARE for the opportunity to join this awesome project.
Footnotes
References
Adams, C. I. M., Knapp, M., Gemmell, N. J., Jeunen, G. J., Bunce, M., Lamare, M. D., et al. (2019). Beyond biodiversity: can environmental DNA (eDNA) cut it as a population genetics tool? Genes 10:192. doi: 10.3390/genes10030192
Ahern, A. L. M., Gomez-Gutierrez, J., Aburto-Oropeza, O., Saldierna-Martinez, R. J., Johnson, A. F., Harada, A. E., et al. (2018). DNA sequencing of fish eggs and larvae reveals high species diversity and seasonal changes in spawning activity in the southeastern Gulf of California. Mar. Ecol. Prog. Ser. 592, 159–179. doi: 10.3354/meps12446
Alberdi, A., Aizpurua, O., Gilbert, M. T. P., and Bohmann, K. (2018). Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 9, 134–147. doi: 10.1111/2041-210x.12849
Aller, E. S. T., Jagd, L. M., Kliebenstein, D. J., and Burow, M. (2018). Comparison of the relative potential for epigenetic and genetic variation to contribute to trait stability. G3 8, 1733–1746. doi: 10.1534/g3.118.200127
Althaus, F., Williams, A., Schlacher, T. A., Kloser, R. J., Green, M. A., Barker, B. A., et al. (2009). Impacts of bottom trawling on deep-coral ecosystems of seamounts are long-lasting. Mar. Ecol. Prog. Ser. 397, 279–294. doi: 10.3354/meps08248
Andruszkiewicz, E. A., Koseff, J. R., Fringer, O. B., Ouellette, N. T., Lowe, A. B., Edwards, C. A., et al. (2019). Modeling environmental DNA transport in the coastal ocean using lagrangian particle tracking. Fron. Mar. Sci. 6:477. doi: 10.3389/fmars.2019.00477
Ardura, A., and Zaiko, A. (2018). PCR-based assay for Mya arenaria detection from marine environmental samples and tracking its invasion in coastal ecosystems. J. Nat. Conserv. 43, 1–7. doi: 10.1016/j.jnc.2018.02.007
Ardura, A., Zaiko, A., Martinez, J. L., Samuiloviene, A., Borrell, Y., and Garcia-Vazquez, E. (2015). Environmental DNA evidence of transfer of North Sea molluscs across tropical waters through ballast water. J. Mollusc. Stud. 81, 495–501. doi: 10.1093/mollus/eyv022
Armbrecht, L. H., Coolen, M. J. L., Lejzerowicz, F., George, S. C., Negandhi, K., Suzuki, Y., et al. (2019). Ancient DNA from marine sediments: precautions and considerations for seafloor coring, sample handling and data generation. Earth Sci. Rev. 196:102887.
Asch, R. G., Stock, C. A., and Sarmiento, J. L. (2019). Climate change impacts on mismatches between phytoplankton blooms and fish spawning phenology. Glob. Chang. Biol. 25, 2544–2559. doi: 10.1111/gcb.14650
Asche, F., and Smith, M. D. (2018). Viewpoint: induced innovation in fisheries and aquaculture. Food Policy 76, 1–7.
Baker, C. S., Steel, D., Nieukirk, S., and Klinck, H. (2018). Environmental DNA (eDNA) from the wake of the whales: droplet digital PCR for detection and species identification. Front. Mar. Sci. 5:133. doi: 10.3389/fmars.2018.00133
Ball, S. L., Hebert, P. D. N., Burian, S. K., and Webb, J. M. (2005). Biological identifications of mayflies (Ephemeroptera) using DNA barcodes. J. N. Am. Benthol. Soc. 24, 508–524. doi: 10.1899/04-142.1
Barnes, M. A., and Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 17, 1–17. doi: 10.1007/s10592-015-0775-4
Batchelder, H. P., Edwards, C. A., and Powell, T. M. (2002). Individual-based models of copepod populations in coastal upwelling regions: implications of physiologically and environmentally influenced diel vertical migration on demographic success and nearshore retention. Prog. Oceanogr. 53, 307–333. doi: 10.1016/S0079-6611(02)00035-6
Beaugrand, G., Mcquatters-Gollop, A., Edwards, M., and Goberville, E. (2012). Long-term responses of North Atlantic calcifying plankton to climate change. Nat. Clim. Chang. 3, 263–267. doi: 10.1038/nclimate1753
Bentur, Y., Ashkar, J., Lurie, Y., Levy, Y., Azzam, Z. S., Litmanovich, M., et al. (2008). Lessepsian migration and tetrodotoxin poisoning due to Lagocephalus sceleratus in the eastern Mediterranean. Toxicon 52, 964–968. doi: 10.1016/j.toxicon.2008.10.001
Berger-Tal, O., Polak, T., Oron, A., Lubin, Y., Kotler, B. P., and Saltz, D. (2011). Integrating animal behavior and conservation biology: a conceptual framework. Behav. Ecol. 22, 236–239. doi: 10.1093/beheco/arq224
Bernstein, A. S., and Ludwig, D. S. (2008). The importance of biodiversity to medicine. J. Am. Med. Assoc. 300, 2297–2299. doi: 10.1001/jama.2008.655
Berry, T. E., Saunders, B. J., Coghlan, M. L., Stat, M., Jarman, S., Richardson, A. J., et al. (2019). Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and identifies ecosystem responses to anomalous climatic events. PLoS Genet. 15:e1007943. doi: 10.1371/journal.pgen.1007943
Bik, H. M., Porazinska, D. L., Creer, S., Caporaso, J. G., Knight, R., and Thomas, W. K. (2012). Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol. Evol. 27, 233–243. doi: 10.1016/j.tree.2011.11.010
Billett, D. S. M., Jones, D. O. B., and Weaver, P. P. E. (2019). “Environmental issues of deep-sea mining,” in Improving Environmental Management Practices in Deep-Sea Mining, ed. R. Sharma, (Cham: Springer International Publishing), 403–446.
Birnbaum, K., Jung, J. W., Wang, J. Y., Lambert, G. M., Hirst, J. A., Galbraith, D. W., et al. (2005). Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619. doi: 10.1038/nmeth0805-615
Block, B. A., Jonsen, I. D., Jorgensen, S. J., Winship, A. J., Shaffer, S. A., Bograd, S. J., et al. (2011). Tracking apex marine predator movements in a dynamic ocean. Nature 475, 86–90. doi: 10.1038/nature10082
Boerder, K., Schiller, L., and Worm, B. (2019). Not all who wander are lost: improving spatial protection for large pelagic fishes. Mar. Policy 105, 80–90. doi: 10.1016/j.marpol.2019.04.013
Bohmann, K., Evans, A., Gilbert, M. T., Carvalho, G. R., Creer, S., Knapp, M., et al. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367. doi: 10.1016/j.tree.2014.04.003
Boussarie, G., Bakker, J., Wangensteen, O. S., Mariani, S., Bonnin, L., Juhel, J.-B., et al. (2018). Environmental DNA illuminates the dark diversity of sharks. Sci. Adv. 4:eaap9661. doi: 10.1126/sciadv.aap9661
Bravington, M. V., Skaug, H. J., and Anderson, E. C. (2016). Close-Kin mark-recapture. Statist. Sci. 31, 259–274. doi: 10.1214/16-sts552
Brechtel, A., Gross, T., and Drossel, B. (2019). Far-ranging generalist top predators enhance the stability of meta-foodwebs. Sci. Rep. 9:12268. doi: 10.1038/s41598-019-48731-y
Brennan, G., Kregting, L., Beatty, G. E., Cole, C., Elsasser, B., Savidge, G., et al. (2014). Understanding macroalgal dispersal in a complex hydrodynamic environment: a combined population genetic and physical modelling approach. J. R. Soc. Interf. 11:20140197. doi: 10.1098/rsif.2014.0197
Brennan, R. S., Garrett, A. D., Huber, K. E., Hargarten, H., and Pespeni, M. H. (2019). Rare genetic variation and balanced polymorphisms are important for survival in global change conditions. Proc. R. Soc. B Biol. Sci. 286:943. doi: 10.1098/rspb.2019.0943
Brown, J. R., Gowen, R. J., and Mclusky, D. S. (1987). The effect of salmon farming on the benthos of a Scottish sea loch. J. Exper. Mar. Biol. Ecol. 109, 39–51. doi: 10.1016/0022-0981(87)90184-5
Buxton, A. S., Groombridge, J. J., Zakaria, N. B., and Griffiths, R. A. (2017). Seasonal variation in environmental DNA in relation to population size and environmental factors. Sci. Rep. 7:46294. doi: 10.1038/srep46294
Bylemans, J., Furlan, E. M., Hardy, C. M., Mcguffie, P., Lintermans, M., and Gleeson, D. M. (2017). An environmental DNA-based method for monitoring spawning activity: a case study, using the endangered Macquarie perch (Macquaria australasica). Methods Ecol. Evol. 8, 646–655. doi: 10.1111/2041-210x.12709
Bylemans, J., Furlan, E. M., Pearce, L., Daly, T., and Gleeson, D. M. (2016). Improving the containment of a freshwater invader using environmental DNA (eDNA) based monitoring. Biol. Invas. 18, 3081–3089. doi: 10.1007/s10530-016-1203-5
Bylemans, J., Gleeson, D. M., Duncan, R. P., Hardy, C. M., and Furlan, E. M. (2019). A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes. Environ. DNA 1, 402–414. doi: 10.1002/edn3.41
Cardinale, B. J., Duffy, J. E., Gonzalez, A., Hooper, D. U., Perrings, C., Venail, P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 486, 59–67. doi: 10.1038/nature11148
Carraro, L., Hartikainen, H., Jokela, J., Bertuzzo, E., and Rinaldo, A. (2018). Estimating species distribution and abundance in river networks using environmental DNA. Proc. Natl. Acad. Sci. U.S.A. 115, 11724–11729. doi: 10.1073/pnas.1813843115
Charlesworth, D., and Charlesworth, B. (1987). Inbreeding depression and its evolutionary cosequences. Annu. Rev. Ecol. Syst. 18, 237–268. doi: 10.1146/annurev.es.18.110187.001321
Charlesworth, D., and Willis, J. H. (2009). The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796. doi: 10.1038/nrg2664
Christensen, K., Doblhammer, G., Rau, R., and Vaupel, J. W. (2009). Ageing populations: the challenges ahead. Lancet 374, 1196–1208.
Clark, S. J., Lee, H. J., Smallwood, S. A., Kelsey, G., and Reik, W. (2016). Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 17:72. doi: 10.1186/s13059-016-0944-x
Collins, R. A., Wangensteen, O. S., O’gorman, E. J., Mariani, S., Sims, D. W., and Genner, M. J. (2018). Persistence of environmental DNA in marine systems. Commun. Biol. 1:185. doi: 10.1038/s42003-018-0192-6
Cordier, T., Frontalini, F., Cermakova, K., Apotheloz-Perret-Gentil, L., Treglia, M., Scantamburlo, E., et al. (2019). Multi-marker eDNA metabarcoding survey to assess the environmental impact of three offshore gas platforms in the North Adriatic Sea (Italy). Mar. Environ. Res. 146, 24–34. doi: 10.1016/j.marenvres.2018.12.009
Costanza, R., D’arge, R., De Groot, R., Farber, S., Grasso, M., Hannon, B., et al. (1997). The value of the world’s ecosystem services and natural capital. Nature 387, 253–260. doi: 10.1038/387253a0
Cowart, D. A., Murphy, K. R., and Cheng, C. H. C. (2018). Metagenomic sequencing of environmental DNA reveals marine faunal assemblages from the West Antarctic Peninsula. Mar. Genom. 37, 148–160. doi: 10.1016/j.margen.2017.11.003
Creer, S., Fonseca, V. G., Porazinska, D. L., Giblin-Davis, R. M., Sung, W., Power, D. M., et al. (2010). Ultrasequencing of the meiofaunal biosphere: practice, pitfalls and promises. Mol. Ecol. 19, 4–20. doi: 10.1111/j.1365-294X.2009.04473.x
Cristescu, M. E. (2019). Can environmental RNA revolutionize biodiversity science? Trends Ecol. Evol. 34, 694–697. doi: 10.1016/j.tree.2019.05.003
Croll, D. A., Baldo, M., Scott, B., Francisco, P. C., Nancy, B., Richard, T., et al. (2005). From wind to whales: trophic links in a coastal upwelling system. Mar. Ecol. Prog. Ser. 289, 117–130.
Crowl, T. A., Crist, T. O., Parmenter, R. R., Belovsky, G., and Lugo, A. E. (2008). The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 6, 238–246. doi: 10.1890/070151
Croxall, J. P., Everson, I., Kooyman, G. L., Ricketts, C., and Davis, R. W. (1985). Fur seal diving behaviour in relation to vertical distribution of krill. J. Anim. Ecol. 54, 1–8. doi: 10.2307/4616
Culhane, F. E., Frid, C. L. J., Royo Gelabert, E., White, L., and Robinson, L. A. (2018). Linking marine ecosystems with the services they supply: what are the relevant service providing units? Ecol. Appl. 28, 1740–1751. doi: 10.1002/eap.1779
Currier, C. A., Morris, T. J., Wilson, C. C., and Freeland, J. R. (2018). Validation of environmental DNA (eDNA) as a detection tool for at-risk freshwater pearly mussel species (Bivalvia: Unionidae). Aquat. Conserv. Mar. Freshw. Ecosyst. 28, 545–558. doi: 10.1002/aqc.2869
Darling, J. A., and Blum, M. J. (2007). DNA-based methods for monitoring invasive species: a review and prospectus. Biol. Invas. 9, 751–765. doi: 10.1007/s10530-006-9079-4
Deegan, L. A. (1993). Nutrient and energy transport between estuaries and coastal marine ecosystems by fish migration. Can. J. Fish. Aquat. Sci. 50, 74–79. doi: 10.1139/f93-009
Dhar, C. B., Cimarelli, L., Singh, S. K., Brandi, L., Brandi, A., Puccinelli, C., et al. (2015). Molecular detection of a potentially toxic diatom species. Intern. J. Environ. Res. Public Health 12, 4921–4941. doi: 10.3390/ijerph120504921
Diederich, S., Nehls, G., Van Beusekom, J. E. E., and Reise, K. (2004). Introduced Pacific oysters (Crassostrea gigas) in the northern Wadden Sea: invasion accelerated by warm summers? Helgoland Mar. Res. 59, 97–106. doi: 10.1007/s10152-004-0195-1
Dodd, L. F., Grabowski, J. H., Piehler, M. F., Westfield, I., and Ries, J. B. (2015). Ocean acidification impairs crab foraging behaviour. Proc. R. Soc. B 282:20150333. doi: 10.1098/rspb.2015.0333
Doney, S. C., Ruckelshaus, M., Duffy, J. E., Barry, J. P., Chan, F., English, C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Dowle, E., Pochon, X., Keeley, N., and Wood, S. A. (2015). Assessing the effects of salmon farming seabed enrichment using bacterial community diversity and high-throughput sequencing. FEMS Microbiol. Ecol. 91:fiv089. doi: 10.1093/femsec/fiv089
Dunn, J., Hall, C. D., Heath, M. R., Mitchell, R. B., and Ritchie, B. J. (1993). ARIES—a system for concurrent physical, biological and chemical sampling at sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 40, 867–878. doi: 10.1016/0967-0637(93)90076-F
Eder, Y., Tschink, D., Gerlach, G., and Strahl, J. (2018). Physiology of juvenile hydroids - High food availability mitigates stress responses of Hydractinia echinata to increasing seawater temperatures. J. Exper. Mar. Biol. Ecol. 508, 64–72. doi: 10.1016/j.jembe.2018.07.009
Edgar, G. J., Stuart-Smith, R. D., Willis, T. J., Kininmonth, S., Baker, S. C., Banks, S., et al. (2014). Global conservation outcomes depend on marine protected areas with five key features. Nature 506, 216–220. doi: 10.1038/nature13022
Edwards, M., and Richardson, A. J. (2004). Impact of climate change on marine pelagic phenology and trophic mismatch. Nature 430, 881–884. doi: 10.1038/nature02808
Egan, S. P., Grey, E., Olds, B., Feder, J. L., Ruggiero, S. T., Tanner, C. E., et al. (2015). Rapid molecular detection of invasive species in ballast and harbor water by integrating environmental DNA and light transmission spectroscopy. Environ. Sci. Technol. 49, 4113–4121. doi: 10.1021/es5058659
Elbrecht, V., Vamos, E. E., Steinke, D., and Leese, F. (2018). Estimating intraspecific genetic diversity from community DNA metabarcoding data. PeerJ 6:e4644. doi: 10.7717/peerj.4644
Ellis, J. I., Fraser, G., and Russell, J. (2012). Discharged drilling waste from oil and gas platforms and its effects on benthic communities. Mar. Ecol. Prog. Ser. 456, 285–302. doi: 10.3354/meps09622
Estoup, A., and Guillemaud, T. (2010). Reconstructing routes of invasion using genetic data: why, how and so what? Mol. Ecol. 19, 4113–4130. doi: 10.1111/j.1365-294X.2010.04773.x
Feinberg, A. P. (2018). The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med. 378, 1323–1334. doi: 10.1056/NEJMra1402513
Fernandes, K., Van Der Heyde, M., Bunce, M., Dixon, K., Harris, R. J., Wardell-Johnson, G., et al. (2018). DNA metabarcoding—a new approach to fauna monitoring in mine site restoration. Restorat. Ecol. 26, 1098–1107. doi: 10.1111/rec.12868
Ficetola, G. F., Miaud, C., Pompanon, F., and Taberlet, P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425. doi: 10.1098/rsbl.2008.0118
Fischer, J. (2013). “Fish identification tools for biodiversity and fisheries assessments: review and guidance for decision-makers,” in FAO Fisheries and Aquaculture Technical Paper, ed. J. Fischer, (Rome: Marine and Inland Fishery Resources Branch FAO Fisheries and Aquaculture Department).
Fraga, M. F., Ballestar, E., Paz, M. F., Ropero, S., Setien, F., Ballestar, M. L., et al. (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U.S.A. 102:10604. doi: 10.1073/pnas.0500398102
Fraser, H. B., Lam, L. L., Neumann, S. M., and Kobor, M. S. (2012). Population-specificity of human DNA methylation. Genome Biol. 13:R8. doi: 10.1186/gb-2012-13-2-r8
Fremier, A. K., Strickler, K. M., Parzych, J., Powers, S., and Goldberg, C. S. (2019). Stream transport and retention of environmental DNA pulse releases in relation to hydrogeomorphic scaling factors. Environ. Sci. Technol. 53, 6640–6649. doi: 10.1021/acs.est.8b06829
Gardner, T. A., Côté, I. M., Gill, J. A., Grant, A., and Watkinson, A. R. (2005). Hurricanes and caribbean coral reefs: impacts, recovery patterns, and role in long-term decline. Ecology 86, 174–184. doi: 10.1890/04-0141
Gokhman, D., Malul, A., and Carmel, L. (2017). Inferring past environments from ancient epigenomes. Mol. Biol. Evol. 34, 2429–2438. doi: 10.1093/molbev/msx211
Gokhman, D., Meshorer, E., and Carmel, L. (2016). Epigenetics: it’s getting old. past meets future in paleoepigenetics. Trends Ecol. Evol. 31, 290–300.
Goodwin, K. D., Thompson, L. R., Duarte, B., Kahlke, T., Thompson, A. R., Marques, J. C., et al. (2017). DNA sequencing as a tool to monitor marine ecological status. Front. Mar. Sci. 4:107. doi: 10.3389/fmars.2017.00107
Gosset, C., Rives, J., and Labonne, J. (2006). Effect of habitat fragmentation on spawning migration of brown trout (Salmo trutta L.). Ecol. Freshw. Fish 15, 247–254. doi: 10.1111/j.1600-0633.2006.00144.x
Graham, S. E., Chariton, A. A., and Landis, W. G. (2019). Using Bayesian networks to predict risk to estuary water quality and patterns of benthic environmental DNA in Queensland. Integrat. Environ. Assess. Manag. 15, 93–111. doi: 10.1002/ieam.4091
Guilfoyle, M. P., and Schultz, M. T. (2017). The contribution of double-crested cormorants (Phalacrocorax auritus) to silver carp (Hypophthalmichthys molitrix) DNA loads in the Chicago Area waterway system. J. Great Lakes Res. 43, 1181–1185.
Guo, H., Zhu, P., Wu, X., Li, X., Wen, L., and Tang, F. (2013). Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 23, 2126–2135. doi: 10.1101/gr.161679.113
Halpern, B. S., Walbridge, S., Selkoe, K. A., Kappel, C. V., Micheli, F., D’agrosa, C., et al. (2008). A global map of human impact on marine ecosystems. Science 319, 948–952. doi: 10.1126/science.1149345
Hänfling, B., Carvalho, G. R., and Brandl, R. (2002). mt-DNA sequences and possible invasion pathways of the Chinese mitten crab. Mar. Ecol. Prog. Ser. 238, 307–310.
Hansen, B. K., Bekkevold, D., Clausen, L. W., and Nielsen, E. E. (2018). The sceptical optimist: challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish. 19, 751–768. doi: 10.1111/faf.12286
Harper, L. R., Handley, L. L., Hahn, C., Boonham, N., Rees, H. C., Gough, K. C., et al. (2018). Needle in a haystack? A comparison of eDNA metabarcoding and targeted qPCR for detection of the great crested newt (Triturus cristatus). Ecol. Evol. 8, 6330–6341. doi: 10.1002/ece3.4013
He, X. P., Sutherland, T. F., Pawlowski, J., and Abbott, C. L. (2019). Responses of foraminifera communities to aquaculture-derived organic enrichment as revealed by environmental DNA metabarcoding. Mol. Ecol. 28, 1138–1153. doi: 10.1111/mec.15007
Hebert, P. D., Cywinska, A., Ball, S. L., and Dewaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Natl. Acad. Sci. U.S.A. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hill, M. O. (1973). Diversity and evenness: a unifying notation and its consequences. Ecology 54, 427–432. doi: 10.2307/1934352
Hillebrand, H., and Cardinale, B. J. (2010). A critique for meta-analyses and the productivity diversity relationship. Ecology 91, 2545–2549.
Hodapp, D., Hillebrand, H., and Striebel, M. (2019). “Unifying” the concept of resource use efficiency in ecology. Front. Ecol. Evol. 6:233. doi: 10.3389/fevo.2018.00233
Hooper, D. U., Chapin, F. S. III, Ewel, J. J., Hector, A., Inchausti, P., Lavorel, S., et al. (2005). Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75, 3–35. doi: 10.1890/04-0922
Hopkins, G. W., and Freckleton, R. P. (2002). Declines in the numbers of amateur and professional taxonomists: implications for conservation. Anim. Conserv. 5, 245–249. doi: 10.1017/s1367943002002299
Howarth, R., Chan, F., Conley, D. J., Garnier, J., Doney, S. C., Marino, R., et al. (2011). Coupled biogeochemical cycles: eutrophication and hypoxia in temperate estuaries and coastal marine ecosystems. Front. Ecol. Environ. 9, 18–26. doi: 10.1890/100008
Hutchings, J. A., Minto, C., Ricard, D., Baum, J. K., and Jensen, O. P. (2010). Trends in the abundance of marine fishes. Can. J. Fish. Aquat. Sci. 67, 1205–1210. doi: 10.1139/F10-081
IPBES, (2019). “Summary for policymakers of the global assessment report on biodiversity and ecosystem services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services,” in Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services, eds S. Díaz, J. Settele, E. S. Brondízio, H. T. Ngo, M. Guèze, J. Agard, et al. (Bonn: IBES secretariat).
IPCC, (2014). “Climate Change 2014: synthesis report. contribution of working groups I, II and III to the fifth assessment report of the intergovernmental panel on climate change,” in Climate Change 2014: Synthesis Report, eds Core Writing Team, R. K. Pachauri and L. A. Meyer, (Geneva: IPCC).
Jensen, O. P., Branch, T. A., and Hilborn, R. (2011). Marine fisheries as ecological experiments. Theoret. Ecol. 5, 3–22. doi: 10.1007/s12080-011-0146-9
Jentsch, A., Kreyling, J., Boettcher-Treschkow, J., and Beierkuhnlein, C. (2009). Beyond gradual warming: extreme weather events alter flower phenology of European grassland and heath species. Glob. Chang. Biol. 15, 837–849. doi: 10.1111/j.1365-2486.2008.01690.x
Jerde, C. L. (2019). Can we manage fisheries with the inherent uncertainty from eDNA? J. Fish Biol. 98, 341–353. doi: 10.1111/jfb.14218
Jo, T., Murakami, H., Yamamoto, S., Masuda, R., and Minamoto, T. (2019). Effect of water temperature and fish biomass on environmental DNA shedding, degradation, and size distribution. Ecol. Evol. 9, 1135–1146. doi: 10.1002/ece3.4802
Kaartvedt, S., Melle, W., Knutsen, T., and Skjoldal, H. R. (1996). Vertical distribution of fish and krill beneath water of varying optical properties. Mar. Ecol. Prog. Ser. 136, 51–58.
Kalantzi, I., and Karakassis, I. (2006). Benthic impacts of fish farming: meta-analysis of community and geochemical data. Mar. Pollut. Bull. 52, 484–493. doi: 10.1016/j.marpolbul.2005.09.034
Kamoroff, C., and Goldberg, C. S. (2018). Environmental DNA quantification in a spatial and temporal context: a case study examining the removal of brook trout from a high alpine basin. Limnology 19, 335–342. doi: 10.1007/s10201-018-0551-5
Katsanevakis, S., Wallentinus, I., Zenetos, A., Leppäkoski, E., Çinar, M. E., Oztürk, B., et al. (2014). Impacts of invasive alien marine species on ecosystem services and biodiversity: a pan-European review. Aquat. Invas. 9, 391–423. doi: 10.3391/ai.2014.9.4.01
Kavanaugh, M. T., Oliver, M. J., Chavez, F. P., Letelier, R. M., Muller-Karger, F. E., and Doney, S. C. (2016). Seascapes as a new vernacular for pelagic ocean monitoring, management and conservation. ICES J. Mar. Sci. 73, 1839–1850. doi: 10.1093/icesjms/fsw086
Kelly, R. P., Gallego, R., and Jacobs-Palmer, E. (2018). The effect of tides on nearshore environmental DNA. PeerJ 6:e4521. doi: 10.7717/peerj.4521
Kelly, R. P., Shelton, A. O., and Gallego, R. (2019). Understanding PCR processes to draw meaningful conclusions from environmental DNA studies. Sci. Rep. 9:12133. doi: 10.1038/s41598-019-48546-x
Kimbro, D. L., Cheng, B. S., and Grosholz, E. D. (2013). Biotic resistance in marine environments. Ecol. Lett. 16, 821–833. doi: 10.1111/ele.12106
Kleinhappel, T. K., Burman, O. H. P., John, E. A., Wilkinson, A., and Pike, T. W. (2019). The impact of water pH on association preferences in fish. Ethology 125, 195–202. doi: 10.1111/eth.12843
Klymus, K. E., Richter, C. A., Chapman, D. C., and Paukert, C. (2015). Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 183, 77–84. doi: 10.1016/j.biocon.2014.11.020
Kochmann, J., Buschbaum, C., Volkenborn, N., and Reise, K. (2008). Shift from native mussels to alien oysters: differential effects of ecosystem engineers. J. Exper. Mar. Biol. Ecol. 364, 1–10.
Krishnamurthy, K. P., and Francis, R. A. (2012). A critical review on the utility of DNA barcoding in biodiversity conservation. Biodivers. Conserv. 21, 1901–1919. doi: 10.1007/s10531-012-0306-2
Kujawinski, E. B. (2011). The impact of microbial metabolism on marine dissolved organic matter. Annu. Rev. Mar. Sci. 3, 567–599. doi: 10.1146/annurev-marine-120308-081003
Lacoursiere-Roussel, A., Cote, G., Leclerc, V., and Bernatchez, L. (2016). Quantifying relative fish abundance with eDNA: a promising tool for fisheries management. J. Appl. Ecol. 53, 1148–1157. doi: 10.1111/1365-2664.12598
Lacoursiere-Roussel, A., Howland, K., Normandeau, E., Grey, E. K., Archambault, P., Deiner, K., et al. (2018). eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol. Evol. 8, 7763–7777. doi: 10.1002/ece3.4213
Lande, R. (1996). Statistics and partitioning of species diversity, and similarity among multiple communities. Oikos 76, 5–13. doi: 10.2307/3545743
Laroche, O., Pochon, X., Tremblay, L. A., Ellis, J. I., Lear, G., and Wood, S. A. (2018). Incorporating molecular-based functional and co-occurrence network properties into benthic marine impact assessments. FEMS Microbiol. Ecol. 94:fiy167. doi: 10.1093/femsec/fiy167
Lascelles, B., Notarbartolo Di Sciara, G., Agardy, T., Cuttelod, A., Eckert, S., Glowka, L., et al. (2014). Migratory marine species: their status, threats and conservation management needs. Aquat. Conserv. Mar. Freshw. Ecosyst. 24, 111–127. doi: 10.1002/aqc.2512
Leis, J. M. (2007). Behaviour as input for modelling dispersal of fish larvae: behaviour, biogeography, hydrodynamics, ontogeny, physiology and phylogeny meet hydrography. Mar. Ecol. Prog. Ser. 347, 185–193. doi: 10.3354/meps06977
Lejzerowicz, F., Esling, P., Pillet, L., Wilding, T. A., Black, K. D., and Pawlowski, J. (2015). High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 5:13932. doi: 10.1038/srep13932
Levi, T., Allen, J. M., Bell, D., Joyce, J., Russell, J. R., Tallmon, D. A., et al. (2019). Environmental DNA for the enumeration and management of Pacific salmon. Mol. Ecol. Resourc. 19, 597–608. doi: 10.1111/1755-0998.12987
Levin, L. A., Ekau, W., Gooday, A. J., Jorissen, F., Middelburg, J. J., Naqvi, S. W. A., et al. (2009). Effects of natural and human-induced hypoxia on coastal benthos. Biogeosciences 6, 2063–2098. doi: 10.5194/bg-6-2063-2009
Litchman, E., and Klausmeier, C. A. (2008). Trait-based community ecology of phytoplankton. Annu. Rev. Ecol. Evol. Syst. 39, 615–639. doi: 10.1146/annurev.ecolsys.39.110707.173549
Liu, W., and He, M. (2012). Effects of ocean acidification on the metabolic rates of three species of bivalve from southern coast of China. Chin. J. Oceanol. Limnol. 30, 206–211. doi: 10.1007/s00343-012-1067-1
Lohrer, A. M., Townsend, M., Rodil, I. F., Hewitt, J. E., and Thrush, S. F. (2012). Detecting shifts in ecosystem functioning: the decoupling of fundamental relationships with increased pollutant stress on sandflats. Mar. Pollut. Bull. 64, 2761–2769.
Lowe, A. T., Roberts, E. A., and Galloway, A. W. E. (2016). Improved marine-derived POM availability and increased pH related to freshwater influence in an inland sea. Limnol. Oceanogr. 61, 2122–2138. doi: 10.1002/lno.10357
Lyons, K. G., Brigham, C. A., Traut, B. H., and Schwartz, M. W. (2005). Rare species and ecosystem functioning. Conserv. Biol. 19, 1019–1024. doi: 10.1111/j.1523-1739.2005.00106.x
Lyons, K. G., and Schwartz, M. W. (2001). Rare species loss alters ecosystem function - invasion resistance. Ecol. Lett. 4, 358–365. doi: 10.1046/j.1461-0248.2001.00235.x
Mace, G. M., Norris, K., and Fitter, A. H. (2012). Biodiversity and ecosystem services: a multilayered relationship. Trends Ecol. Evol. 27, 19–26. doi: 10.1016/j.tree.2011.08.006
Mächler, E., Osathanunkul, M., and Altermatt, F. (2018). Shedding light on eDNA: neither natural levels of UV radiation nor the presence of a filter feeder affect eDNA-based detection of aquatic organisms. PLoS One 13:e0195529. doi: 10.1371/journal.pone.0195529
MacKenzie, D. I., and Bailey, L. L. (2004). Assessing the fit of site-occupancy models. J. Agric. Biol. Environ. Statist. 9, 300–318. doi: 10.1198/108571104X3361
Mariani, S., Baillie, C., Colosimo, G., and Riesgo, A. (2019). Sponges as natural environmental DNA samplers. Curr. Biol. 29, R401–R402. doi: 10.1016/j.cub.2019.04.031
Markert, A., Wehrmann, A., and Kröncke, I. (2009). Recently established Crassostrea-reefs versus native Mytilus-beds: differences in ecosystem engineering affects the macrofaunal communities (Wadden Sea of Lower Saxony, southern German Bight). Biol. Invas. 12, 15–32. doi: 10.1007/s10530-009-9425-4
Marques, T. A., Thomas, L., Martin, S. W., Mellinger, D. K., Ward, J. A., Moretti, D. J., et al. (2013). Estimating animal population density using passive acoustics. Biol. Rev. 88, 287–309. doi: 10.1111/brv.12001
Marshall, N. T., and Stepien, C. A. (2019). Invasion genetics from eDNA and thousands of larvae: a targeted metabarcoding assay that distinguishes species and population variation of zebra and quagga mussels. Ecol. Evol. 9, 3515–3538. doi: 10.1002/ece3.4985
Martínez, M. L., Intralawan, A., Vázquez, G., Pérez-Maqueo, O., Sutton, P., and Landgrave, R. (2007). The coasts of our world: ecological, economic and social importance. Ecol. Econ. 63, 254–272. doi: 10.1016/j.marpolbul.2005.09.034
Mauvisseau, Q., Burian, A., Gibson, C., Brys, R., Ramsey, A., and Sweet, M. (2019). Influence of accuracy, repeatability and detection probability in the reliability of species-specific eDNA based approaches. Sci. Rep. 9:580. doi: 10.1038/s41598-018-37001-y
McClanahan, T. R., Marnane, M. J., Cinner, J. E., and Kiene, W. E. (2006). A comparison of marine protected areas and alternative approaches to coral-reef management. Curr. Biol. 16, 1408–1413. doi: 10.1016/j.cub.2006.05.062
McGill, B. J., Dornelas, M., Gotelli, N. J., and Magurran, A. E. (2015). Fifteen forms of biodiversity trend in the Anthropocene. Trends Ecol. Evol. 30, 104–113. doi: 10.1016/j.tree.2014.11.006
McManus, G. B., and Katz, L. A. (2009). Molecular and morphological methods for identifying plankton: what makes a successful marriage? J. Plankton Res. 31, 1119–1129. doi: 10.1093/plankt/fbp061
Merder, J., Browne, P., Freund, J. A., Fullbrook, L., Graham, C., Johnson, M. P., et al. (2020). Density-dependent growth in ‘catch-and-wait’ fisheries has implications for fisheries management and Marine Protected Areas. Ambio 49, 107–117. doi: 10.1007/s13280-019-01158-1
Molnar, J. L., Gamboa, R. L., Revenga, C., and Spalding, M. D. (2008). Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ. 6, 485–492. doi: 10.1890/070064
Mostofa, K. M. G., Liu, C. Q., Zhai, W., Minella, M., Vione, D., Gao, K., et al. (2016). Reviews and syntheses: ocean acidification and its potential impacts on marine ecosystems. Biogeosciences 13, 1767–1786. doi: 10.5194/bg-13-1767-2016
Mouillot, D., Bellwood, D. R., Baraloto, C., Chave, J., Galzin, R., Harmelin-Vivien, M., et al. (2013). Rare species support vulnerable functions in high-diversity ecosystems. PLoS Biol. 11:e1001569. doi: 10.1371/journal.pbio.1001569
Mouquet, N., Devictor, V., Meynard, C. N., Munoz, F., Bersier, L.-F., Chave, J., et al. (2012). Ecophylogenetics: advances and perspectives. Biol. Rev. 87, 769–785. doi: 10.1111/j.1469-185X.2012.00224.x
Murakami, H., Yoon, S., Kasai, A., Minamoto, T., Yamamoto, S., Sakata, M. K., et al. (2019). Dispersion and degradation of environmental DNA from caged fish in a marine environment. Fish. Sci. 85:1109. doi: 10.1007/s12562-019-01341-z
Nagelkerken, I., and Munday, P. L. (2016). Animal behaviour shapes the ecological effects of ocean acidification and warming: moving from individual to community-level responses. Glob. Chang. Biol. 22, 974–989. doi: 10.1111/gcb.13167
Nätt, D., Rubin, C.-J., Wright, D., Johnsson, M., Beltéky, J., Andersson, L., et al. (2012). Heritable genome-wide variation of gene expression and promoter methylation between wild and domesticated chickens. BMC Genom. 13:59. doi: 10.1186/1471-2164-13-59
Norris, K. (2004). Managing threatened species: the ecological toolbox, evolutionary theory and declining-population paradigm. J. Appl. Ecol. 41, 413–426.
North, E. W., Schlag, Z., Hood, R. R., Li, M., Zhong, L., Gross, T., et al. (2008). Vertical swimming behavior influences the dispersal of simulated oyster larvae in a coupled particle-tracking and hydrodynamic model of Chesapeake Bay. Mar. Ecol. Prog. Ser. 359, 99–115. doi: 10.3354/meps07317
Occhipinti-Ambrogi, A., and Galil, B. (2010). Marine alien species as an aspect of global change. Adv. Oceanogr. Limnol. 1, 199–218. doi: 10.1080/19475721003743876
O’Donnell, J. L., Kelly, R. P., Shelton, A. O., Samhouri, J. F., Lowell, N. C., and Williams, G. D. (2017). Spatial distribution of environmental DNA in a nearshore marine habitat. PeerJ 5:e3044. doi: 10.7717/peerj.3044
Olsgard, F., Somerfield, P. J., and Carr, M. R. (1998). Relationships between taxonomic resolution, macrobenthic community patterns and disturbance. Mar. Ecol. Prog. Ser. 172, 25–36.
Ore, J.-P., Elbaum, S., Burgin, A., and Detweiler, C. (2015). Autonomous aerial water sampling. J. Field Robot. 32, 1095–1113. doi: 10.1002/rob.21591
Osterholz, H., Singer, G., Wemheuer, B., Daniel, R., Simon, M., Niggemann, J., et al. (2016). Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 10:1717. doi: 10.1038/ismej.2015.231
Pan, K., and Wang, W.-X. (2012). Trace metal contamination in estuarine and coastal environments in China. Sci. Total Environ. 421–422, 3–16. doi: 10.1016/j.scitotenv.2011.03.013
Parsons, K. M., Everett, M., Dahlheim, M., and Park, L. (2018). Water, water everywhere: environmental DNA can unlock population structure in elusive marine species. R. Soc. Open Sci. 5:180537. doi: 10.1098/rsos.180537
Pawlowski, J., Esling, P., Lejzerowicz, F., Cedhagen, T., and Wilding, T. A. (2014). Environmental monitoring through protist next-generation sequencing metabarcoding: assessing the impact of fish farming on benthic foraminifera communities. Mol. Ecol. Resour. 14, 1129–1140. doi: 10.1111/1755-0998.12261
Pawlowski, J., Kelly-Quinn, M., Altermatt, F., Apothéloz-Perret-Gentil, L., Beja, P., Boggero, A., et al. (2018). The future of biotic indices in the ecogenomic era: integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 637–638, 1295–1310. doi: 10.1016/j.scitotenv.2018.05.002
Pejchar, L., and Mooney, H. A. (2009). Invasive species, ecosystem services and human well-being. Trends Ecol. Evol. 24, 497–504. doi: 10.1016/j.tree.2009.03.016
Phillips, A. (2004). The history of the international system of protected area management categories. Intern. J. Protect. Area Manag. 14, 4–14.
Pielou, E. C. (1966). The measurement of diversity in different types of biological collections. J. Theoret. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Pikitch, E. K., Santora, C., Babcock, E. A., Bakun, A., Bonfil, R., Conover, D. O., et al. (2004). Ecosystem-based fishery management. Science 305, 346–347.
Pirotta, V., Smith, A., Ostrowski, M., Russell, D., Jonsen, I. D., Grech, A., et al. (2017). An economical custom-built drone for assessing whale health. Front. Mar. Sci. 4:425. doi: 10.3389/fmars.2017.00425
Pochon, X., Wood, S. A., Keeley, N. B., Lejzerowicz, F., Esling, P., Drew, J., et al. (2015). Accurate assessment of the impact of salmon farming on benthic sediment enrichment using foraminiferal metabarcoding. Mar. Pollut. Bull. 100, 370–382. doi: 10.1016/j.marpolbul.2015.08.022
Pochon, X., Zaiko, A., Fletcher, L. M., Laroche, O., and Wood, S. A. (2017). Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLoS One 12:e0187636. doi: 10.1371/journal.pone.0187636
Pomati, F., Jokela, J., Simona, M., Veronesi, M., and Ibelings, B. W. (2011). An automated platform for phytoplankton ecology and aquatic ecosystem monitoring. Environ. Sci. Technol. 45, 9658–9665. doi: 10.1021/es201934n
Pomerantz, A., Peñafiel, N., Arteaga, A., Bustamante, L., Pichardo, F., Coloma, L. A., et al. (2018). Real-time DNA barcoding in a rainforest using nanopore sequencing: opportunities for rapid biodiversity assessments and local capacity building. Gigascience 7:giy033. doi: 10.1093/gigascience/giy033
Por, F. D. (1971). One hundred years of suez canal—A century of lessepsian migration: retrospect and viewpoints. Syst. Biol. 20, 138–159. doi: 10.2307/2412054
Prendergast, J. R., Quinn, R. M., Lawton, J. H., Eversham, B. C., and Gibbons, D. W. (1993). Rare species, the coincidence of diversity hotspots and conservation strategies. Nature 365, 335–337. doi: 10.1038/365335a0
Preston, C. M., Harris, A., Ryan, J. P., Roman, B., Marin, R. III, Jensen, S., et al. (2011). Underwater application of quantitative pcr on an ocean mooring. PLoS One 6:e22522. doi: 10.1371/journal.pone.0022522
Ptacnik, R., Solimini, A. G., Andersen, T., Tamminen, T., Brettum, P., Lepisto, L., et al. (2008). Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc. Natl. Acad. Sci. U.S.A. 105, 5134–5138. doi: 10.1073/pnas.0708328105
Rabalais, N. N. (2002). Nitrogen in aquatic ecosystems. Ambio 31, 102–112. doi: 10.1579/0044-7447-31.2.102
Radulovici, A. E., Sainte-Marie, B., and Dufresne, F. (2009). DNA barcoding of marine crustaceans from the Estuary and Gulf of St lawrence: a regional-scale approach. Mol. Ecol. Resour. 9, 181–187. doi: 10.1111/j.1755-0998.2009.02643.x
Rakyan, V. K., Blewitt, M. E., Druker, R., Preis, J. I., and Whitelaw, E. (2002). Metastable epialleles in mammals. Trends Genet. 18, 348–351.
Reise, K., Buschbaum, C., Büttger, H., and Wegner, K. M. (2017). Invading oysters and native mussels: from hostile takeover to compatible bedfellows. Ecosphere 8:e01949. doi: 10.1002/ecs2.1949
Reusch, T. B., Ehlers, A., Hammerli, A., and Worm, B. (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl. Acad. Sci. U.S.A. 102, 2826–2831. doi: 10.1073/pnas.0500008102
Robinson, C. V., De Leaniz, C. G., and Consuegra, S. (2019). Effect of artificial barriers on the distribution of the invasive signal crayfish and Chinese mitten crab. Sci. Rep. 9:11. doi: 10.1038/s41598-019-43570-3
Roman, J., Estes, J. A., Morissette, L., Smith, C., Costa, D., Mccarthy, J., et al. (2014). Whales as marine ecosystem engineers. Front. Ecol. Environ. 12, 377–385. doi: 10.1890/130220
Runge, C. A., Watson, J. E. M., Butchart, S. H. M., Hanson, J. O., Possingham, H. P., and Fuller, R. A. (2015). Protected areas and global conservation of migratory birds. Science 350, 1255–1258. doi: 10.1126/science.aac9180
Ruppert, K. M., Kline, R. J., and Rahman, M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17:e00547. doi: 10.1016/j.gecco.2019.e00547
Ryther, J. H. (1969). Photosynthesis and fish production in the sea. Science 166:72. doi: 10.1126/science.166.3901.72
Sacco, M., Blyth, A., Bateman, P. W., Hua, Q., Mazumder, D., Nicole, W., et al. (2019). New light in the dark - a proposed multidisciplinary framework for studying functional ecology of groundwater fauna. Sci. Total Environ. 662, 963–977. doi: 10.1016/j.scitotenv.2019.01.296
Sahu, R., Srivastava, P. C., and Kota, V. K. B. (2013). Deformed shell model results for neutrinoless positron double beta decay of nuclei in theA= 60-90 region. J. Phys. G Nuclear Particle Phys. 40:095107. doi: 10.1088/0954-3899/40/9/095107
Salter, I. (2018). Seasonal variability in the persistence of dissolved environmental DNA (eDNA) in a marine system: the role of microbial nutrient limitation. PLoS One 13:e0192409. doi: 10.1371/journal.pone.0192409
Salter, I., Joensen, M., Kristiansen, R., Steingrund, P., and Vestergaard, P. (2019). Environmental DNA concentrations are correlated with regional biomass of Atlantic cod in oceanic waters. Commun. Biol. 2:461. doi: 10.1038/s42003-019-0696-8
Sassoubre, L. M., Yamahara, K. M., Gardner, L. D., Block, B. A., and Boehm, A. B. (2016). Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ. Sci. Technol. 50, 10456–10464. doi: 10.1021/acs.est.6b03114
Schlötterer, C., Tobler, R., Kofler, R., and Nolte, V. (2014). Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15, 749–763. doi: 10.1038/nrg3803
Schmelzle, M. C., and Kinziger, A. P. (2016). Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Mol. Ecol. Resour. 16, 895–908. doi: 10.1111/1755-0998.12501
Schmidt, B. R., Kéry, M., Ursenbacher, S., Hyman, O. J., Collins, J. P., and Yoccoz, N. (2013). Site occupancy models in the analysis of environmental DNA presence/absence surveys: a case study of an emerging amphibian pathogen. Methods Ecol. Evol. 4, 646–653. doi: 10.1111/2041-210x.12052
Scholin, C. A. (2010). What are “ecogenomic sensors?” A review and thoughts for the future. Ocean Sci. 6, 51–60. doi: 10.5194/os-6-51-2010
Segner, H., Caroll, K., Fenske, M., Janssen, C. R., Maack, G., Pascoe, D., et al. (2003). Identification of endocrine-disrupting effects in aquatic vertebrates and invertebrates: report from the European IDEA project. Ecotoxicol. Environ. Saf. 54, 302–314. doi: 10.1016/S0147-6513(02)00039-8
Siegenthaler, A., Wangensteen, O. S., Soto, A. Z., Benvenuto, C., Corrigan, L., and Mariani, S. (2019). Metabarcoding of shrimp stomach content: harnessing a natural sampler for fish biodiversity monitoring. Mol. Ecol. Resour. 19, 206–220. doi: 10.1111/1755-0998.12956
Sigsgaard, E. E., Jensen, M. R., Winkelmann, I. E., Møller, P. R., Hansen, M. M., and Thomsen, P. F. (2020). Population-level inferences from environmental DNA—Current status and future perspectives. Evol. Appl. 13, 245–262. doi: 10.1111/eva.12882
Sigsgaard, E. E., Nielsen, I. B., Carl, H., Krag, M. A., Knudsen, S. W., Xing, Y. C., et al. (2017). Seawater environmental DNA reflects seasonality of a coastal fish community. Mar. Biol. 164:128. doi: 10.1007/s00227-017-3147-4
Skliris, N., and Djenidi, S. (2006). Plankton dynamics controlled by hydrodynamic processes near a submarine canyon off NW corsican coast: a numerical modelling study. Continent. Shelf Res. 26, 1336–1358. doi: 10.1016/j.csr.2006.05.004
Smallwood, S. A., Lee, H. J., Angermueller, C., Krueger, F., Saadeh, H., Peat, J., et al. (2014). Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820. doi: 10.1038/nmeth.3035
Smart, A. S., Tingley, R., Weeks, A. R., Van Rooyen, A. R., and Mccarthy, M. A. (2015). Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecol. Appl. 25, 1944–1952. doi: 10.1890/14-1751.1
Smart, A. S., Weeks, A. R., Van Rooyen, A. R., Moore, A., Mccarthy, M. A., and Tingley, R. (2016). Assessing the cost-efficiency of environmental DNA sampling. Methods Ecol. Evol. 7, 1291–1298. doi: 10.1111/2041-210x.12598
Smith, C. J., and Ryckman, K. K. (2015). Epigenetic and developmental influences on the risk of obesity, diabetes, and metabolic syndrome. Diabetes 8, 295–302. doi: 10.2147/DMSO.S61296
Spear, S. F., Groves, J. D., Williams, L. A., and Waits, L. P. (2015). Using environmental DNA methods to improve detectability in a hellbender (Cryptobranchus alleganiensis) monitoring program. Biol. Conserv. 183, 38–45. doi: 10.1016/j.biocon.2014.11.016
Stat, M., Huggett, M. J., Bernasconi, R., Dibattista, J. D., Berry, T. E., Newman, S. J., et al. (2017). Ecosystem biomonitoring with eDNA: metabarcoding across the tree of life in a tropical marine environment. Sci. Rep. 7:12240. doi: 10.1038/s41598-017-12501-5
Stat, M., John, J., Dibattista, J. D., Newman, S. J., Bunce, M., and Harvey, E. S. (2019). Combined use of eDNA metabarcoding and video surveillance for the assessment of fish biodiversity. Conserv. Biol. 33, 196–205. doi: 10.1111/cobi.13183
Stern, R. F., Picard, K. T., Hamilton, K. M., Walne, A., Tarran, G. A., Mills, D., et al. (2015). Novel lineage patterns from an automated water sampler to probe marine microbial biodiversity with ships of opportunity. Prog. Oceanogr. 137, 409–420. doi: 10.1016/j.pocean.2015.04.015
Stoeck, T., Frühe, L., Forster, D., Cordier, T., Martins, C. I. M., and Pawlowski, J. (2018a). Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture. Mar. Pollut. Bull. 127, 139–149. doi: 10.1016/j.marpolbul.2017.11.065
Stoeck, T., Kochems, R., Forster, D., Lejzerowicz, F., and Pawlowski, J. (2018b). Metabarcoding of benthic ciliate communities shows high potential for environmental monitoring in salmon aquaculture. Ecol. Indic. 85, 153–164. doi: 10.1016/j.ecolind.2017.10.041
Stoeckle, M. Y., Soboleva, L., and Charlop-Powers, Z. (2017). Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary. PLoS One 12:e0175186. doi: 10.1371/journal.pone.0175186
Strickler, K. M., Fremier, A. K., and Goldberg, C. S. (2015). Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 183, 85–92. doi: 10.1016/j.biocon.2014.11.038
Stuckas, H., Knobel, L., Schade, H., Breusing, C., Hinrichsen, H. H., Bartel, M., et al. (2017). Combining hydrodynamic modelling with genetics: can passive larval drift shape the genetic structure of Baltic Mytilus populations? Mol. Ecol. 26, 2765–2782. doi: 10.1111/mec.14075
Taberlet, P., Bonin, A., Zinger, L., and Coissac, E. (2018). Environmental DNA: For Biodiversity Research and Monitoring. Oxford: Oxford University Press.
Taberlet, P., Coissac, E., Hajibabae, M., and Rieseberg, L. H. (2012). Environmental DNA. Mol. Ecol. 21, 1789–1793.
Taylor, H. R., and Harris, W. E. (2012). An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Mol. Ecol. Resour. 12, 377–388. doi: 10.1111/j.1755-0998.2012.03119.x
Thomsen, P. F., Kielgast, J., Iversen, L. L., Moller, P. R., Rasmussen, M., and Willerslev, E. (2012a). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS One 7:e41732. doi: 10.1371/journal.pone.0041732
Thomsen, P. F., Kielgast, J., Iversen, L. L., Wiuf, C., Rasmussen, M., Gilbert, M. T., et al. (2012b). Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 21, 2565–2573. doi: 10.1111/j.1365-294X.2011.05418.x
Thomsen, P. F., Moller, P. R., Sigsgaard, E. E., Knudsen, S. W., Jorgensen, O. A., and Willerslev, E. (2016). Environmental dna from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS One 11:e0165252. doi: 10.1371/journal.pone.0165252
Thresher, R. E., and Kuris, A. M. (2004). Options for managing invasive marine species. Biol. Invas. 6, 295–300.
Truelove, N. K., Andruszkiewicz, E. A., and Block, B. A. (2019). A rapid environmental DNA method for detecting white sharks in the open ocean. Methods Ecol. Evol. 10, 1128–1135. doi: 10.1111/2041-210x.13201
Tucker, C. M., Cadotte, M. W., Carvalho, S. B., Davies, T. J., Ferrier, S., Fritz, S. A., et al. (2017). A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 92, 698–715. doi: 10.1111/brv.12252
Turner, C. R., Uy, K. L., and Everhart, R. C. (2015). Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol. Conserv. 183, 93–102. doi: 10.1016/j.biocon.2014.11.017
Turon, X., Antich, A., Palacín, C., Praebel, K., and Wangensteen, O. S. (2019). From metabarcoding to metaphylogeography: separating the wheat from the chaff. Ecol. Appl. 30:e02036. doi: 10.1002/eap.2036
Ussler, W., Preston, C., Tavormina, P., Pargett, D., Jensen, S., Roman, B., et al. (2013). Autonomous application of quantitative PCR in the deep sea: in situ surveys of aerobic methanotrophs using the deep-sea environmental sample processor. Environ. Sci. Technol. 47, 9339–9346. doi: 10.1021/es4023199
Uthicke, S., Lamare, M., and Doyle, J. R. (2018). eDNA detection of corallivorous seastar (Acanthaster cf. solaris) outbreaks on the great barrier reef using digital droplet PCR. Coral Reefs 37, 1229–1239. doi: 10.1007/s00338-018-1734-6
Valentini, A., Taberlet, P., Miaud, C., Civade, R., Herder, J., Thomsen, P. F., et al. (2016). Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 25, 929–942. doi: 10.1111/mec.13428
Weithoff, G., and Beisner, B. E. (2019). Measures and approaches in trait-based phytoplankton community ecology - from freshwater to marine ecosystems. Front. Mar. Sci. 6:40. doi: 10.3389/fmars.2019.00040
Wilcox, T. M., Mckelvey, K. S., Young, M. K., Sepulveda, A. J., Shepard, B. B., Jane, S. F., et al. (2016). Understanding environmental DNA detection probabilities: a case study using a stream-dwelling char Salvelinus fontinalis. Biol. Conserv. 194, 209–216. doi: 10.1016/j.biocon.2015.12.023
Wilcox, T. M., Young, M. K., Mckelvey, K. S., Isaak, D. J., Horan, D. L., and Schwartz, M. K. (2018). Fine-scale environmental DNA sampling reveals climate-mediated interactions between native and invasive trout species. Ecosphere 9:e02500. doi: 10.1002/ecs2.2500
Worm, B., Barbier, E. B., Beaumont, N., Duffy, J. E., Folke, C., Halpern, B. S., et al. (2006). Impacts of biodiversity loss on ocean ecosystem services. Science 314, 787–790. doi: 10.1126/science.1132294
Worm, B., Hilborn, R., Baum, J. K., Branch, T. A., Collie, J. S., Costello, C., et al. (2009). Rebuilding global fisheries. Science 325, 578–585. doi: 10.1126/science.1173146
Xie, Y., Zhang, X., Yang, J., Kim, S., Hong, S., Giesy, J. P., et al. (2018). eDNA-based bioassessment of coastal sediments impacted by an oil spill. Environ. Pollut. 238, 739–748. doi: 10.1016/j.envpol.2018.02.081
Xu, N., Zhu, B., Shi, F., Shao, K., Que, Y. F., Li, W. T., et al. (2018). Monitoring seasonal distribution of an endangered anadromous sturgeon in a large river using environmental DNA. Sci. Nat. 105:62. doi: 10.1007/s00114-018-1587-4
Yamahara, K. M., Preston, C. M., Birch, J., Walz, K., Marin, R., Jensen, S., et al. (2019). In situ autonomous acquisition and preservation of marine environmental DNA Using an autonomous underwater vehicle. Front. Mar. Sci. 6:373. doi: 10.3389/fmars.2019.00373
Yamamoto, S., Masuda, R., Sato, Y., Sado, T., Araki, H., Kondoh, M., et al. (2017). Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 7:40368. doi: 10.1038/srep40368
Yang, C.-H., and Andrew Pospisilik, J. (2019). Polyphenism - A window into gene-environment interactions and phenotypic plasticity. Front. Genet. 10:132. doi: 10.3389/fgene.2019.00132
Yates, M. C., Fraser, D. J., and Derry, A. M. (2019). Meta-analysis supports further refinement of eDNA for monitoring aquatic species-specific abundance in nature. Environ. DNA 1, 5–13. doi: 10.1002/edn3.7
Zaiko, A., Martinez, J. L., Schmidt-Petersen, J., Ribicic, D., Samuiloviene, A., and Garcia-Vazquez, E. (2015a). Metabarcoding approach for the ballast water surveillance - An advantageous solution or an awkward challenge? Mar. Pollut. Bull. 92, 25–34. doi: 10.1016/j.marpolbul.2015.01.008
Zaiko, A., Samuiloviene, A., Ardura, A., and Garcia-Vazquez, E. (2015b). Metabarcoding approach for nonindigenous species surveillance in marine coastal waters. Mar. Pollut. Bull. 100, 53–59. doi: 10.1016/j.marpolbul.2015.09.030
Zaiko, A., Pochon, X., Garcia-Vazquez, E., Olenin, S., and Wood, S. A. (2018). Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: management and surveillance of non-indigenous species. Front. Mar. Sci. 5:322. doi: 10.3389/fmars.2018.00322
Zhang, X. W. (2019). Environmental DNA shaping a new era of ecotoxicological research. Environ. Sci. Technol. 53, 5605–5612. doi: 10.1021/acs.est.8b06631
Zhao, L., Liu, D., Xu, J., Wang, Z., Chen, Y., Lei, C., et al. (2018). The framework for population epigenetic study. Brief. Bioinform. 19:bbw098.
Keywords: environmental DNA, eDNA, fisheries management, marine conservation, population genetics, ecosystem functioning
Citation: Schadewell Y and Adams CIM (2021) Forensics Meets Ecology – Environmental DNA Offers New Capabilities for Marine Ecosystem and Fisheries Research. Front. Mar. Sci. 8:668822. doi: 10.3389/fmars.2021.668822
Received: 17 February 2021; Accepted: 26 March 2021;
Published: 27 April 2021.
Edited by:
Simon Jungblut, University of Bremen, GermanyReviewed by:
Rupert Collins, University of Bristol, United KingdomNaiara Guimaraes Sales, University of Salford, United Kingdom
Copyright © 2021 Schadewell and Adams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yvonne Schadewell, eXZvbm5lLnNjaGFkZXdlbGxAdW9sLmRl
 Yvonne Schadewell
Yvonne Schadewell Clare I. M. Adams
Clare I. M. Adams