Museum Genomics Illuminate the High Specificity of a Bioluminescent Symbiosis for a Genus of Reef Fish
- California Academy of Sciences, San Francisco, CA, United States
Symbiotic relationships between bioluminescent bacteria and fishes have evolved multiple times across hundreds of fish taxa, but relatively little is known about the specificity of these associations and how stable they are over host generations. This study describes the degree of specificity of a bioluminescent symbiosis between cardinalfishes in the genus Siphamia and luminous bacteria in the Vibrio family. Primarily using museum specimens, we investigated the codivergence of host and symbiont and test for patterns of divergence that correlate with both biogeography and time. Contrary to expectations, we determined that the light organ symbionts of all 14 Siphamia species examined belong to one genetic clade of Photobacterium mandapamensis (Clade II), indicating that the association is highly specific and conserved throughout the host genus. Thus, we did not find evidence of codivergence among hosts and symbionts. We did observe that symbionts hosted by individuals sampled from colder water regions were more divergent, containing more than three times as many single nucleotide polymorphisms than the rest of the symbionts examined. Overall, our findings indicate that the symbiosis between Siphamia fishes and P. mandapamensis Clade II has been highly conserved across host taxa and over a broad geographic range despite the facultative nature of the bacterial symbiont. We also present a new approach to simultaneously recover genetic information from a bacterial symbiont and its vertebrate host from formalin-fixed specimens, enhancing the utility of museum collections.
Introduction
Bioluminescent symbioses have evolved multiple times across diverse fish and squid taxa, including at least 17 times in the ray-finned fishes (Dunlap and Urbanczyk, 2013; Davis et al., 2016). Approximately 500 species of fish are known to be symbiotically bioluminescent, but the range of luminous bacteria they associate with is much narrower than what would be predicted based solely on the diversity and relative abundances of bacteria in the surrounding seawater. For example, both leiognathid fishes and anglerfishes representing four different families have each been described to host only two species of closely related luminous bacteria (Reichelt et al., 1977; Kaeding et al., 2007; Baker et al., 2019). A similar level of bacterial specificity has been described in at least seven additional fish families with symbiotically luminous members (Dunlap et al., 2007). Despite this seemingly high level of specificity for environmentally transmitted symbioses, our understanding of these associations is lacking, as the majority have yet to be characterized. Moreover, luminous bacterial symbionts are generally not obligately dependent on their host (but see Hendry et al., 2014, 2018) and can survive in a variety of other habitats including seawater, sediment, and the surfaces and digestive tracts of various marine organisms. Thus, these facultative symbionts must retain the genetic machinery necessary to associate with their hosts while also being able to compete with the rest of the microbial community in the environment (Bright and Bulgheresi, 2010), bringing up questions of the evolutionary history of these associations and the mechanisms that maintain specificity.
Luminous bacteria involved in bioluminescent symbioses are members of the Vibrionaceae family, and of these, three species of Photobacterium are known to associate with fish hosts: P. kishitanii, P. leiognathi, and P. mandapamensis. Photobacterium leiognathi and P. mandapamensis are closely related and phenotypically similar and were once considered to be the same species based on the inability of 16S rRNA and gyrB genes to distinguish between them (Ast and Dunlap, 2004). Ecologically, however, symbiotic strains of P. leiognathi and P. mandapamensis are distinct; P. mandapamensis has a broad host range and has been identified from the light organs of fishes from at least four families representing two teleost orders collected from diverse marine habitats and depths, while P. leiognathi primarily associates with various leiognathid fishes (Kaeding et al., 2007). Furthermore, an analysis of the lux genes, which are responsible for light production, indicated that luxF is present in P. mandapamensis yet absent in P. leiognathi. Thus, these two groups were considered to be distinct subspecies of P. leiognathi (Ast and Dunlap, 2004). An additional analysis of various strains of P. mandapamensis and P. leiognathi isolated from fish light organs further discriminated P. mandapamensis from P. leiognathi and revealed two distinct clades within P. mandapamensis (Clades I and II) based on gyrB and luxAB(F)E genes (Kaeding et al., 2007), although the physiological and ecological significance of these two clades remain unknown. In that study, a high degree of specificity was also observed between the the cardinalfish host Siphamia tubifer and Clade II of P. mandapamensis.
Bioluminescence has evolved multiple times within the cardinalfish family (Perciformes: Apogonidae), but only the genus Siphamia rely on a symbiotic relationship with bacteria to produce light; other cardinalfishes produce their own light presumably via the acquisition of luciferin from their diet (Thacker and Roje, 2009). Siphamia consists of 25 species all of which are symbiotically bioluminescent (Thacker and Roje, 2009; Gon and Allen, 2012). Siphamia fishes possess a ventral light organ connected to the intestine, which contains a dense population of luminous bacteria (~108 cells) (Dunlap and Nakamura, 2011). The light organ symbionts of only one Siphamia species, S. tubifer, originating from a small region in the Okinawa Islands, Japan, have been characterized to date; of the hundreds of specimens examined, all hosted members of Clade II of P. mandapamensis in their light organs, suggesting a high level of specificity for this association (Kaeding et al., 2007; Gould and Dunlap, 2019). However, S. tubifer is broadly distributed throughout the Indo-Pacific, spanning from eastern Africa to the French Polynesian Islands (Gon and Allen, 2012), thus the degree of specificity of the association across its broad geographic range remains unknown. Furthermore, the luminous symbionts of the other 24 Siphamia species have yet to be identified.
The primary goals of this study were to characterize the degree of specificity of the bioluminescent symbiosis throughout the Siphamia genus and across the broad geographic range of S. tubifer. More specifically, we wanted to determine whether the high degree of specificity observed for S. tubifer and members of Clade II of Photobacterium mandpamensis (Kaeding et al., 2007) is maintained across the host genus, and if not, whether any observed symbiont diversity is a result of codivergence between host and symbiont. By leveraging readily available museum specimens from several natural history collections we were able to sample geographically and temporally diverse Siphamia taxa to address these questions (Figure 1). However, recovering genetic information from specimens in wet collections, particularly those initially fixed in formalin, is challenging primarily due to DNA degradation and cross-linkage. Here we present a new approach to simultaneously recover genetic information from a bacterial symbiont and its vertebrate host from formalin-fixed specimens based on a combination of recently developed methods. This allowed us to sample diverse Siphamia taxa spanning decades throughout the Indo-Pacific as well as S. tubifer from locations throughout its entire geographic range (Figure 1). With these samples we were able to test for patterns of symbiont diversity at the subspecies level that correlate with host biogeography, temperature and time. Although codivergence is unlikely to occur when the symbiont has a free-living stage (Ronquist, 1998) and has not been documented for other symbiotically bioluminescent fishes (Dunlap et al., 2007), our ability to recover informative genetic information from both the host and symbiont also allowed us to test for evidence of codivergence in this bioluminescent association.
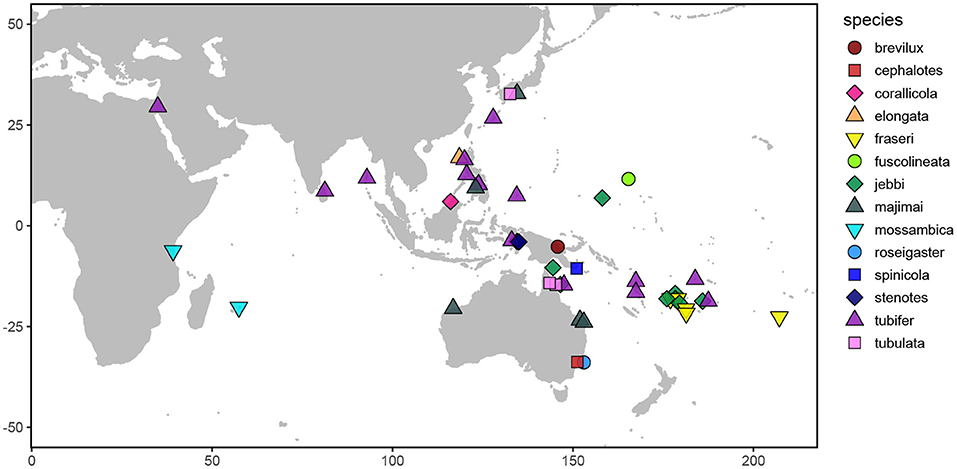
Figure 1. Map depicting the sampling locations of the Siphamia specimens examined in this study. Distinct colors and shapes represent the different Siphamia species sampled as indicated in the figure legend.
Methods
Taxon Sampling and DNA Extraction
We sampled 59 specimens representing 14 Siphamia species obtained from the combined wet collections of the California Academy of Sciences, the Australian Museum, and the Smithsonian National Museum of Natural History, and including several newly collected specimens deposited at the California Academy of Sciences (Figures 1, 2, Table 1). The specimens collected for this study were stored in 96% ethanol and were obtained with appropriate collection permits, following approved protocols and permits for the capture, care and handling of fish by the California Academy of Science's Institutional Animal Care and Use Committee. To extract DNA from each specimen, we adapted the following protocol from two previous methods designed for recovering DAN from formalin-fixed tissues (Hykin et al., 2015; Ruane and Austin, 2017). Light organs were aseptically dissected and individually placed into 1 ml of GTE buffer and allowed to soak for 3 h at room temperature. This step was repeated two times, after which each light organ was transferred into a final 1 ml aliquot of fresh GTE buffer and left to soak overnight at room temperature. The following morning, each sample was transferred into 1 ml of 100% ethanol for 1 min, followed by 1 ml of 70% ethanol for 5 min, and 1 ml of nuclease-free water for 10 min at room temperature. Light organs were then transferred into 180 ul of pre-heated (98°C) ATL buffer (QIAGEN) and incubated at 98°C for 15 min, after which samples were immediately placed on ice for at least 2 min. Once cooled, 40 ul of proteinase K was added to each sample, and the samples were incubated at 60°C for 48 h on a shaking heat block. Samples were vortexed periodically and additional 20 ul aliquots of proteinase K were added as needed (up to 100 ul total). Following this incubation period, DNA was extracted using the QIAGEN DNEasy Blood and Tissue Kit as described by the manufacturer. Purified DNA products were eluted into 50 ul of nuclease-free water after a 3-min incubation at 55°C.
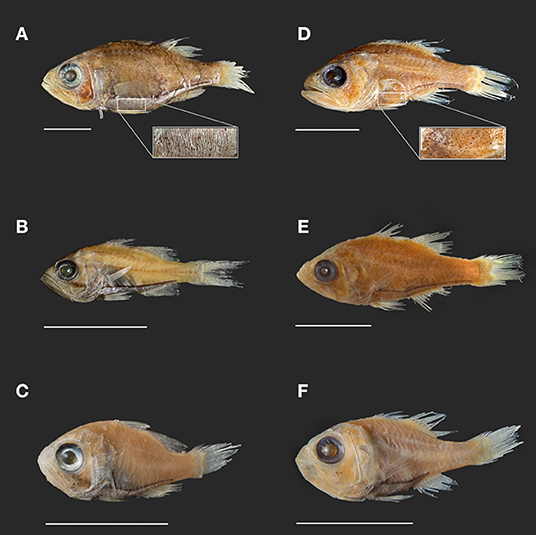
Figure 2. Photographs of select Siphamia specimens from lots used in this study. Specimens (A–C) represent the tubifer subgroup (Gon and Allen, 2012), identified by the striated light organ (A) and specimens (D–F) represent the tubulata subgroup, identified by the spotted light organ (D). (A) S. tubifer (USNM341595) with insert of light organ detail showing striated morphology. (B) S. stenotes (USNM396981, paratype). (C) S. jebbi (CAS223855). (D) S. tubulata (CAS28515) with insert of light organ detail showing spotted morphology. (E) S. corallicola (USNM203781). (F) S. brevilux (CAS65338, paratype). Scale bars indicate 1 cm in length.
Library Preparation and Sequencing
Samples were quantified using the Qubit dsDNA HS Assay Kit on the Qubit 2.0 Fluorometer (Invitrogen) and profiled with an Agilent 2100 Bioanalyzer. Samples with a peak in size distribution >300 bp were sonicated with a Qsonica (Q800R3) for one or two minutes (if peak was >1,500 bp) with a pulse rate of 10–10 s and an amplitude of 25%. Samples were then treated with the NEBNext FFPE DNA Repair Mix following the manufacturer's instructions and DNA libraries were immediately prepped using the NEBNext Ultra II DNA Library Prep Kit. Samples with low or undetectable quantities of dsDNA were re-quantified using the Qubit ssDNA HS Assay Kit and prepared using the Accel-NGS 1S Plus DNA Library Kit (Swift Biosciences), which uses both single- and double-stranded DNA as templates. Each sample was uniquely indexed with the NEBNext Multiplex Oligos for Illumina. Final libraries were cleaned with AMPure XP magnetic beads, pooled, and sequenced as single-end 150 bp (UC Berkeley, QB3) or paired-end 150 bp reads on the Illumina HiSeq 4000 platform (NovoGene), or as paired-end 150 bp reads on the Illumina NovaSeq S4 platform (Genewiz). Supplementary Table 1 contains details for each sample and library preparation.
Sequence Analysis
Sequences were demultiplexed, trimmed and quality filtered for a Phred score of 20 or above using Trimmomatic (Bolger et al., 2014). The remaining reads were aligned to the reference genome of Photobacterium mandapamensis, isolated from the light organ of Siphamia tubifer (Urbanczyk et al., 2011) with BWA-MEM using the default parameters (Li, 2013). Unaligned sequences were then processed with MitoFinder (Allio et al., 2020) using the reference mitochondrial genome of the Banggai cardinalfish Pterapogon kauderni (Matias and Hereward, 2018). All cardinalfish cytochrome oxidase subunit 1 (COI) gene sequences that were recovered were aligned using MUSCLE (Edgar, 2004) and a maximum likelihood analysis was carried out with raxml-ng (Kozlovet al., 2019) using the evolutionary model TIM2+F+I+G4, which had the lowest BIC score as predicted by IQtree (Nguyen et al., 2015), and 1,000 bootstraps to infer the phylogenetic relationships between host species. COI sequences of Siphamia spp. from previous studies were also included in the analysis (Supplementary Table 2). An additional phylogeny was inferred from a supermatrix of 15 mitochondrial genes (ATP6, ATP8, COXI, COX2, COX3, CYTB, ND1, ND2, ND3, ND4, ND4L, ND5, ND6, 16S, 18S) identified by MitoFinder that were present in at least 70% of the individuals included in the analysis using the SuperCRUNCH python toolkit (Portik and Wiens, 2020). The concatenated supermatrix alignment was used in a maximum likelihood analysis by raxml-ng with 500 bootstrap replicates and the evolutionary model TIM2+F+R4 as predicted by IQtree to infer the phylogenetic relationships between species.
For the aligned symbiont reads, two approaches were used to determine symbiont identity based on consensus sequences generated for each sample using the samtools mpileup command and the bcftools consensus caller (Li et al., 2009). First, the program fastANI (Jain et al., 2018) was used to calculate the whole-genome average nucleotide identity (ANI) of each sample relative to several available reference genomes of Photobacterium species previously isolated from fish light organs, P. kishitanii pjapo1.1, P. leiognathi lrivu4.1, P. mandapamensis ajapo4.1 (Clade I), P. mandapamensis gjord1.1 (Clade II), and P. mandapamensis svers1.1 (Clade II). The output represents the percent ANI of all orthologous genes shared between each sample's consensus genome and the reference genomes. We also extracted 16S rRNA gene sequences from each sample by aligning all filtered light organ sequences to the complete 16S sequence of a free-living strain of Photobacterium leiognathi (AY292917) (Nishiguchi and Nair, 2003). We chose this strain of P. leiognathi to capture a range of potential luminous bacteria that could be present in the light organ. A sequence similarity search was then performed with the basic local alignment search tool (BLAST) (Altschul et al., 1990) against NCBI's microbial database to identify the known sequence with the lowest E-value and highest percent identity.
To infer the phylogenetic relationships between symbionts from different hosts, all sequences that aligned to the reference genome of P. mandapamensis (Urbanczyk et al., 2011) were also analyzed for sequence variation with the program snippy (Seemann, 2015). Locus filters required a minimum depth of 10x and presence in at least 90% of all reads to minimize calling errors due to the degraded nature of many museum samples. In snippy, single nucleotide polymorphisms (SNPs) were called by Freebayes, and consensus SNP haplotypes were generated for each sample, presumably representing the most abundant symbiont. A sequence alignment of a core set of these SNPs was then created across all samples, filtering for those that had enough genome coverage to produce a core set of at least 1,000 SNPs. SNPs were also called for two additional reference genomes of P. mandapamensis representing both Clade I (ajapo4.1) and Clade II (Res4.1) and were included in the core set of SNPs as phylogenetic references. The phylogenetic relationships of these bacteria were then inferred with raxml-ng (Kozlov et al., 2019) using the evolutionary model TVM+F+R3, which had the lowest BIC score as predicted by IQtree (Nguyen et al., 2015), and 1,500 bootstrap replicates.
Samples included in both the host and symbiont phylogenies were then compared and tested for co-divergence using the cospeciation function in the R phytools package (Revell, 2011). SNPs were annotated with the program SNPeff (Cingolani et al., 2012). Pairwise phylogenetic (patristic) distances between symbionts were calculated with the adephylo package (Jombart et al., 2010) in R, and pairwise geographic distances were calculated based on each specimen's latitude and longitude using the R package geodist (Padgham and Sumner, 2020). Tests for correlations between the phylogenetic distances for each pair of symbionts and their geographic distance or difference between sampling years were carried out, and P-values were adjusted for multiple comparisons with the Holm method in R.
Results
DNA Recovery
Variable amounts of total DNA were recovered from the light organs of preserved Siphamia specimens, ranging from undetectable levels (<2 ng) to more than 1,500 ng. Although light organ size (~1–3 mm diameter) increases linearly with fish standard length (Gould et al., 2016), there was no correlation between DNA yield and specimen size (Spearman's rank correlation: rho=0.52, P = 0.09). Despite this variability in yield, quality DNA sequences were recovered from several specimens with undetectable levels of starting DNA. In fact, some samples with undetectable levels of input DNA resulted in >90% coverage of the symbiont genome at 10x depth. Of note, many of those sequence libraries were prepared using the Swift Bioscience Accel-NGS 1S Plus DNA Library Kit which uses both double and single stranded DNA as the starting template (Supplementary Table 1).
Host Phylogeny
Host COI sequences were recovered from 32 samples and analyzed with an additional 12 Siphamia COI sequences from previous studies (Supplementary Table 2) to generate a maximum likelihood phylogeny of 17 Siphamia species (Figure 3). The supermatrix of 15 mitochondrial genes from 27 Siphamia specimens representing 11 species resulted in a phylogenetic tree with similar, but not identical, topology, and stronger bootstrap support at the nodes (Supplementary Figure 1).
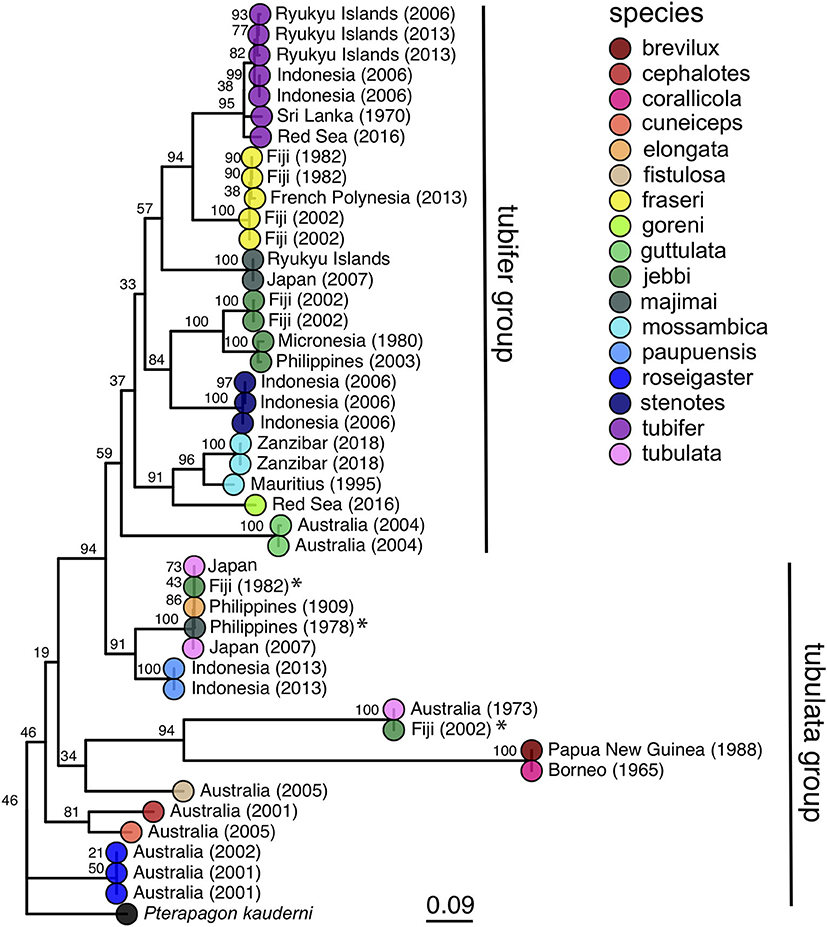
Figure 3. Maximum likelihood phylogeny of cardinalfishes in the genus Siphamia based on COI gene sequences. Species identities are indicated by the branch tip colors and the sampling location and year of each specimen is listed in the branch label. Bootstrap support values are indicated at each node. The Banggai cardinalfish, Pterapogon kauderni, was used as the outgroup. The tubifer and tubulata subgroups within Siphamia (Gon and Allen, 2012) are highlighted with vertical lines to the right of the tree. Specimens that fall outside of their designated subgroup based on species identities are indicated with an *.
Our phylogenetic hypothesis for Siphamia is very similar to that proposed by Gon and Allen (2012) using morphological characters, with slight variations in the placement of specific taxa. Our tree contains a clade that corresponds to Gon and Allen's S. tubifer species group, characterized by a striated pattern on the light organ (Figure 2), although one individual S. majimai and two S. jebbi specimens fell out of this group (Figure 3). Within this group, our trees support the relationships of S. jebbi and S. stenotes as sister species, as well as S. tubifer and S. fraseri. The relative placement of S. mossambica, S. majimai, and S. goreni varies among the trees, but there is support for S. mossambica and S. goreni as sister species in the COI tree, S. mossambica and S. majimai as sisters in the supermatrix tree, and S. majimai and S. goreni as sisters in the morphological tree (Gon and Allen, 2012). As such, it is likely that all of three of these species belong to one clade. The relationships among the species outside of the S. tubifer group are less certain, with several species clustering into species complexes. However, S. roseigaster, S. cuneiceps, and S. cephalotes consistently fall out near the base of the tree, indicating that these species diverged earlier.
Symbiont Identification and Phylogeny
To identify the most abundant light organ symbiont of each Siphamia host, we first calculated the ANI of each sample's consensus symbiont genome relative to the reference genomes of several symbiotic strains of Photobacterium sp. isolated from fish light organs. Eighty-six percent of all the symbionts in this study had ANI values of 95% or greater relative to P. mandapamensis strains in Clade II (gjord1.1 and svers1.1) (Table 2), which is the recommended value to delimit bacterial species (Goris et al., 2007). All remaining samples also had highest ANI values relative to P. mandapamensis Clade II than to the other Photobacterium reference strains, with the exception of one sample (AMI18740-066), which was most similar to P. mandapamensis Clade I (86.8% ANI vs. 85.4% ANI to Clade II) but had fewer orthologous matches (41.7% to Clade I vs. 62.3% to Clade II). None of the symbionts had higher ANI values relative to P. leiognathi than to P. mandapamensis strains. However, many samples had low overall genome coverage, and thus, the ANI calculations are based on only small numbers of orthologous regions (Table 2). To confirm the identities of the light organ symbionts, we also recovered 16S rRNA sequences from the shotgun sequence data and matched them against the NCBI database. Ninety three percent of all samples had >95% coverage of the 16S rRNA gene at 10x read depth. Of these, 67% returned P. mandapamensis as their top hit, and all others returned P. leiognathi (Supplementary Table 3). Importantly, 16S rRNA gene sequences alone are insufficient at discriminating between strains of P. mandapamensis and P. leiognathi (Ast and Dunlap, 2004; Wada et al., 2006). No other Photobacterium species were identified as top hits in this analysis.
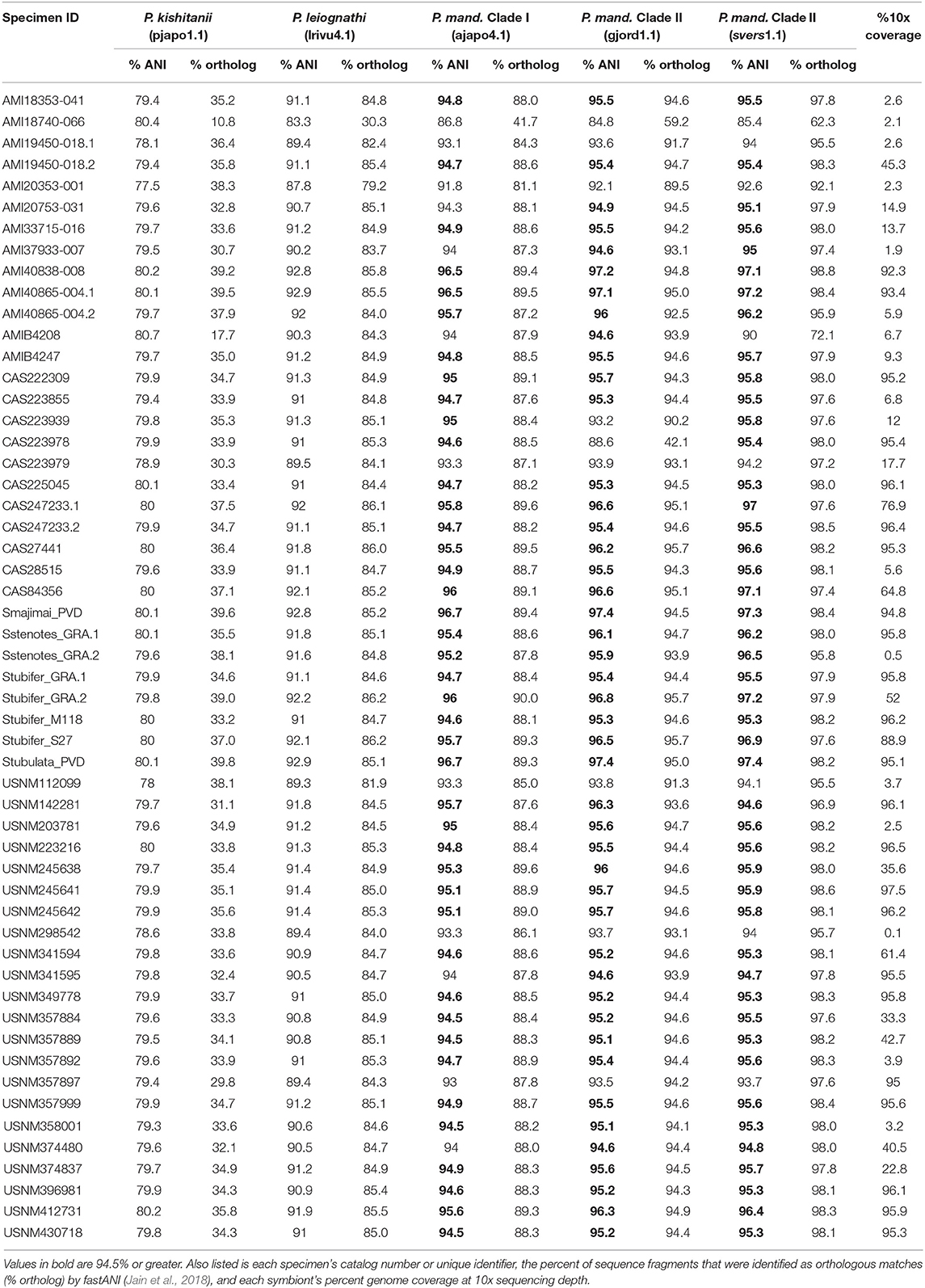
Table 2. Average nucleotide identities (% ANI) of the consensus sequences of the light organ symbionts from the Siphamia specimens sampled in this study relative to several symbiotic Photobacterium species: P. kishitanii (pjapo1.1), P. leiognathi (lrivu4.1), P. mandapamensis (P. mand.), Clade I (ajapo4.1), and Clade II (gjord1.1, svers1.1).
Single nucleotide polymorphisms (SNPs) were detected for the light organ symbionts from most of the specimens sampled, but this number varied greatly and correlated with the variability in genome coverage (Spearman's rank correlation: rho = 0.84, P < 0.001, Supplementary Table 1). Samples with >50% symbiont genome coverage at 10x read depth had an average of 23,221 SNPs relative to the reference genome of P. mandapamensis (Urbanczyk et al., 2011). A core set of 1,471 SNPs were identified across 32 specimens that represent 11 Siphamia host species and included reference genomes from both Clade I and Clade II of Photobacterium mandapamensis. Sixty-eight percent of these SNPs were synonymous, and the remaining non-synonymous SNPs were found in 288 distinct genes. None of the core SNPs were located in the lux operon, composed of the genes responsible for light production. However, two non-synonymous SNPs were detected in the rpoN gene, which is known to play a role in biofilm formation, bioluminescence, and symbiosis initiation for Aliivibrio fischeri (Wolfe et al., 2004), the luminous symbiont of many squid and other fish species. No other SNPs were detected in genes of known function for the bioluminescent symbiosis between A. fischeri and the squid host Euprymna scolopes (Norsworthy and Visick, 2013).
A maximum likelihood phylogeny was inferred for the bacterial symbionts using full sequence alignments that included the core set of SNPs described above. This analysis confirmed that all Siphamia light organ symbionts for which we recovered enough quality sequence data to analyze belong to Clade II of P. mandapamensis and that the reference strain of P. mandapamensis representing Clade I (ajapo4.1) was a clear outgroup (Figure 4). The majority of symbionts analyzed were closely related to the reference strain svers1.1 of P. mandapamensis, although several symbionts fell out in a group with P. mandapamensis strain Res 4.1, both of which are members of Clade II. There were three additional symbionts, all from different host species, that did not belong to either of these subgroups, but are still clearly members of Clade II. We carried out an additional analysis that also included four individuals initially excluded from the analysis because they produced a smaller set of core SNPs (N = 166). The topology of the resulting phylogeny did not change, and these additional symbionts also belong to Clade II of P. mandapamensis (Supplementary Figure 2).
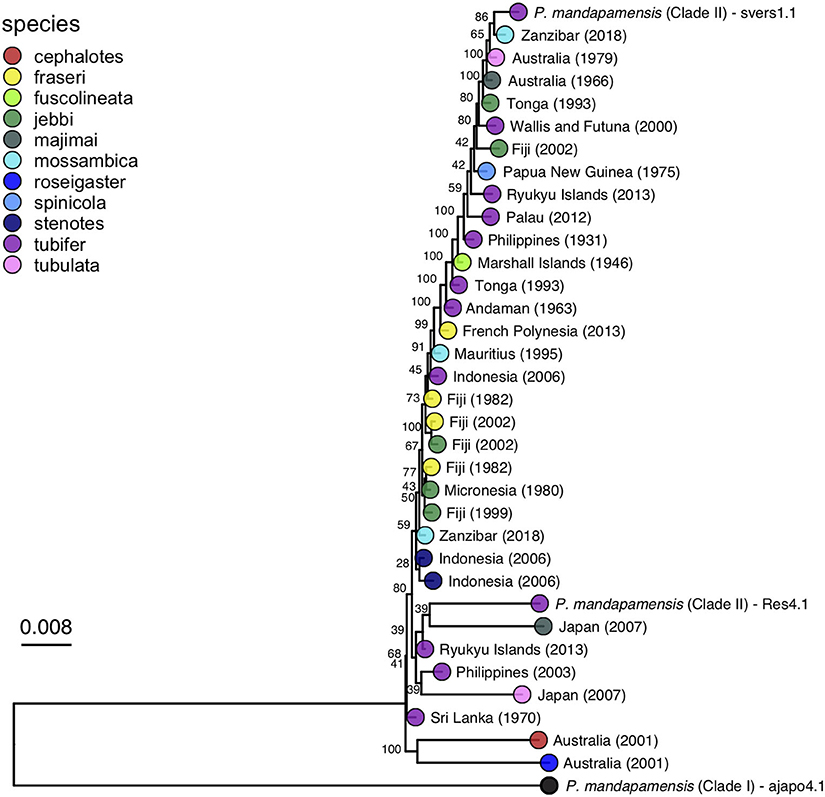
Figure 4. Maximum likelihood phylogeny of the light organ symbionts of various Siphamia species constructed from a core set of 1,471 single nucleotide polymorphisms. Corresponding host species are indicated by the branch tip colors and the sampling location and year of each specimen is listed in the branch label. Bootstrap support values are indicated at each node.
No clear patterns of symbiont divergence that corresponded with host species, geography, or time emerged. There was no correlation between symbiont phylogenetic distance and geographic distance (Spearman's rank correlation: rho=-0.013, Pcorr=1) and there was a slightly negative correlation between phylogenetic distance and time in years (Spearman's rank correlation: rho = −0.17, Pcorr=0.006). In fact, the oldest specimen for which informative sequence data was retained was collected in 1931 and it had luminous bacteria in its light organ that was highly similar to symbionts from specimens collected more than 80 years later. Similarly, Siphamia specimens collected from locations in the western Indian Ocean had symbionts that were closely related to those from locations as far east as Fiji and French Polynesia. With respect to S. tubifer, which has the broadest geographic distribution of all Siphamia species, the symbionts of all ten specimens included in the symbiont phylogeny fell out in Clade II of P. mandapamensis and showed no pattern of strain diversity by geography, confirming the high degree of specificity of this association, even across S. tubifer's broad geographic range.
The bacterial symbionts from four distinct host species had notably longer branches than the others, two of which were closely related to reference strain Res 4.1, an isolate from the light organ of S. tubifer collected in Okinawa, Japan in 2014. Corresponding with longer branch lengths, these four symbionts had more than 3 times as many SNPs than any other sample, ranging between 66,583 and 72,219 SNPs (Supplementary Table 1). Interestingly, these four specimens were collected from two locations, Sydney, Australia and Kochi, Japan, which had the lowest minimum annual temperatures of all collection sites in this study (Table 1). Furthermore, there were 20,082 SNPs in common among these samples that were not present in the core set of SNPs identified across all samples.
Analysis of Co-divergence
Twenty specimens had informative sequence information for both the host and symbiont, and thus, we were able to carry out an analysis of co-divergence based on the host COI phylogeny and corresponding symbiont phylogeny for these individuals. This analysis revealed no evidence of co-divergence of Siphamia hosts and their light organ symbionts (P = 0.13) as seen in Figure 5. However, S. roseigaster and S. cephalotes fall out as sister lineages relative to the rest of Siphamia, and their symbionts follow a similar pattern, forming a sister clade to the rest of P. mandapamensis Clade II.

Figure 5. Analysis of the phylogenetic relationships of Siphamia hosts (left) and their light organ symbionts (right) revealed no evidence of co-divergence. The host cladogram is based on COI gene sequences and the symbiont cladogram is based on a core set of 1,471 single nucleotide polymorphisms. Linkages between individual hosts and their symbionts are shown and colored according to host species.
Discussion
Our results indicate that the symbiosis between cardinalfishes in the genus Siphamia and the luminous bacterium Photobacterium mandapamensis is highly conserved across host species, over geographic space, and over decades. All of the symbionts for which we recovered enough sequence data to analyze were identified as members of Clade II of P. mandapamensis (Kaeding et al., 2007). The subspecies level of symbiont specificity observed between Siphamia species and P. mandapamensis Clade II over the broad geographic and temporal ranges examined is surprising, especially given that the bacteria is a facultative symbiont. Such a high degree of specificity is expected for vertically transmitted symbioses in which a host transfers its symbiotic bacteria directly to its offspring (Moran, 2006), but is unexpected for environmentally transmitted symbioses where symbionts are acquired from a genetically diverse, free-living population of bacteria by each new host generation. This degree of specificity across the Siphamia genus is higher than has been documented for other symbiotically luminous fishes; other genera typically associate with a few different species of luminous bacteria, and two symbiont species have even been described cohabiting within the same light organ (Kaeding et al., 2007). Similarly, within genera of sepiolid and loliginid squids, including Euprymna, hosts associate with several different species of bioluminescent bacteria (Guerrero-Ferreira and Nishiguchi, 2007), further highlighting the markedly higher subspecies-level of symbiont specificity observed throughout the Siphamia genus. This highly conserved relationship between Siphamia and P. mandapamensis Clade II suggests that there are attributes of this clade of bacteria that are essential for the symbiosis as well as selection mechanisms that help each new host generation maintain specificity.
Siphamia tubifer larvae only take up symbionts in their pelagic phase, at least 1 week after hatching, when their light organ becomes receptive to colonization (Dunlap et al., 2012). Yet, Photobacterium spp. normally occurs in relatively low concentrations in the pelagic environment (Troussellier et al., 2017), and even more so at the sub-species level. For a larval host to rely on this improbable encounter in the open water would be considered a very risky strategy. However, it has been shown that S. tubifer hosts regularly excrete their luminous symbiont with fecal waste (Dunlap and Nakamura, 2011), thereby enriching its population in the immediate environment. Indeed, a previous study of S. tubifer symbiont genomics revealed fine-scale population structure of P. mandapamensis Clade II among geographic locations, indicating that symbiont populations are heavily influenced by their local hosts (Gould and Dunlap, 2019), a mechanism that has also been documented for the bioluminescent squid Euprymna scolopes and its luminous symbiont, Aliivibrio fischeri (Lee and Ruby, 1994; Wollenberg and Ruby, 2009), although squid hosts can acquire a luminous symbiont immediately upon hatching (Nyholm and McFall-Ngai, 2004). The specific timing of symbiont acquisition for S. tubifer larvae in the wild has yet to be defined, but this local enrichment may be a key factor in mitigating the risk of relying on environmental transmission of a narrow range of luminous bacteria, and for Siphamia hosts, ensures that P. mandapamensis Clade II will be readily available to new recruits anywhere that populations of Siphamia already occur.
The apparent preference to associate with P. mandapamensis Clade II over strains in Clade I also suggests that there are critical strain-level differences between members of these clades that may be of consequence to the host. However, most studies of microbial symbioses overlook subspecies-level symbiont variation, even though this variation can have important impacts on a host. For example, patterns of strain variation in A. fischeri have been observed within and between populations of their host squid, E. scolopes (Jones et al., 2006; Wollenberg and Ruby, 2009), and different strains have different colonization efficiencies (Lee and Ruby, 1994; Bongrand et al., 2016), mechanisms of biofilm formation during host colonization (Rotman et al., 2019), and could have variable fitness consequences to their host (Koch et al., 2014). In this study we characterized strain variation in P. mandapamensis associated with various Siphamia hosts but saw no distinct correlation between symbiont strain and host species with respect to time or geography. However, we did observe some strain divergence associated with colder temperatures. Four of the Siphamia specimens examined had more than three times as many symbiont SNPs as the others. These four individuals were all collected from more temperate regions in Japan and Australia, which had the lowest minimum annual temperatures of all locations surveyed (Table 1). Temperature is a driving factor of the distribution of bacteria in the marine environment (Sul et al., 2013), and has been shown to influence the symbiotic associations of other marine taxa, such as sepiolid squids (Nishiguchi, 2000; Zamborsky and Nishiguchi, 2011) and cnidarians (Herrera et al., 2020). Thus, the symbionts associated with these four specimens could have genetic adaptations to slightly cooler temperatures. Future studies investigating the influence of temperature on strain diversity and host colonization efficiency would help to elucidate the role that temperature might play in the Siphamia-Photobacterium mandapamensis symbiosis.
Our primary objective of this study was to sequence the symbionts found in the light organs of various Siphamia species, but we were able to recover enough host sequence data to also construct a reasonably well-supported host phylogeny. This allowed us to examine codivergence of hosts and their microbial symbionts. Although we found no evidence of codivergence, the high degree of specificity maintained for this symbiosis across host species over a broad geographic range suggests that this association is genetically constrained by the host. This host-mediated selection poses the question of whether strains in Clade II of P. mandapamensis provide a fitness advantage to the host compared to other bacteria moving through the gut of Siphamia, including P. mandapamensis Clade I and other luminous bacteria. Understanding the genetic architecture of this symbiont selection mechanism may be key to deciphering the highly specific nature of the association. It should be noted that a lack of co-diversification does not preclude a history of coevolution of host and symbiont in the system (Moran, 2006), and members of Clade II of P. mandapamensis are likely to have specific adaptations that provide them with a fitness advantage inside the light organ environment of Siphamia hosts.
Siphamia, the only symbiotically luminous genus of cardinalfish, is monophyletic and divergent from the rest of the Apogonidae (Thacker and Roje, 2009). The absence of this symbiosis in all other cardinalfish genera, including the other bioluminescent genera, brings up intriguing questions regarding the role of the symbiosis in the evolution of the Siphamia genus, specifically whether the ancestral Siphamia evolved via speciation by symbiosis (Wallin, 1927), and whether this association has since acted as a key innovation for the genus. Key innovations help clades persist and diversify by shifting them into a new adaptive zone, increasing fitness, or increasing the propensity for specialization (Heard and Hauser, 1995). In Siphamia, the association with luminous bacteria is thought to provide a fitness benefit either by protecting against predation via counter-illumination or by attracting prey while foraging at night. This fitness advantage could have helped Siphamia persist over evolutionary time, allowing it to diversify. Symbioses have been credited for catalyzing or enhancing adaptive radiations in several systems, such as symbioses with photosynthetic protists in foraminifera (Norris, 1996) and heterobranchia (Wägele, 2004), and symbioses between gall-forming insects and fungi (Joy, 2013).
This study also highlights the potential for formalin-fixed, fluid-preserved museum specimens to be used to investigate microbial symbioses. Adapting recently-developed molecular techniques to extract and prepare DNA from these specimens for sequencing, including the use of single-stranded DNA as templates to construct sequence libraries, we recovered informative sequence data for both the host and its bacterial symbiont. This method allowed us to identify and compare strain-level differences between the bacterial symbionts of many host species collected across dates spanning nearly a century and dispersed widely throughout the Indo-Pacific. However, a limitation of working with formalin-fixed museum specimens is the fragmented nature of the resulting sequence data, which required us to use consensus sequences to characterize the light organ symbionts. Thus, we recognize that our methods were not designed to detect rare symbiont strains. Additional strains, including other Photobacterium species, may have also been present at low abundance but could not be characterized with this approach. However, we are confident that P. mandapamensis Clade II is the most abundant symbiont in all of our Siphamia specimens, and if any alternative symbionts had a comparable abundance, they would have been detected by our SNP analysis and fallen outside of P. mandapamensis Clade II in the phylogenetic tree. Additional investigations using fresh specimens and culture-based methods would be helpful to corroborate our results. Notably, we also saw no clear correlation between sequence quality or yield and variables such as specimen age, size, or DNA input. Initial specimen preservation methods and long-term storage conditions likely contributed more to the variability in DNA quality (Hykin et al., 2015), and this specimen metadata is often lacking. Nevertheless, we were successful at retrieving genetic information from decades old specimens initially fixed in formalin that enabled us to characterize the specificity of the bioluminescent symbiosis across the Siphamia genus throughout the Indo-Pacific. With the continued advancement of genomic techniques and sequencing technologies, our ability to retrieve genetic information for both hosts and bacterial symbionts from museum specimens will further our understanding of these critical associations.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SAMN17043373-SAMN17043448.
Ethics Statement
The animal study was reviewed and approved by California Academy of Sciences IACUC.
Author Contributions
ALG conceived of the project and secured funding for the work. AF-P and ALG carried out the genomic methods and analyses. AG assisted with the identification of specimens and analysis of their associated metadata. All authors contributed to the discussion and interpretation of the results and writing of the manuscript. All authors approve of the submitted version of this manuscript.
Funding
Funding was provided in part by a National Science Foundation Postdoctoral Research Fellowship in Biology (NSF-DBI-1711430) and by the National Institutes for Health (NIH-DP5-OD026405-01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Athena Lam and California Academy of Science's Center for Comparative Genomics for technical support with our genomic methods as well as Joe Russack and James Henderson for their bioinformatics help and advice. Thank you to Luiz Rocha and Hudson Pinheiro for their collection efforts, Dave Catania and Mysi Hoang for their support with the museum specimens, and Jessica Herbert for her assistance in the lab. We also thank the Australian Museum and the Smithsonian National Museum of Natural History for access to their specimens for this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.630207/full#supplementary-material
References
Allio, R., Schomaker-Bastos, A., Romiguier, J., Prosdocimi, F., Nabholz, B., and Delsuc, F. (2020). MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol. Ecol. Resour. 20, 892–905. doi: 10.1111/1755-0998.13160
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ast, J. C., and Dunlap, P. V. (2004). Phylogenetic analysis of the lux operon distinguishes two evolutionarily distinct clades of Photobacterium leiognathi. Arch. Microbiol. 181, 352–361. doi: 10.1007/s00203-004-0663-7
Baker, L. J., Freed, L. L., Easson, C. G., Lopez, J. V., Fenolio, D., Sutton, T. T., et al. (2019). Diverse deep-sea anglerfishes share a genetically reduced luminous symbiont that is acquired from the environment. eLife. 8:e47606. doi: 10.7554/eLife.47606
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bongrand C, Koch, E. J., Moriano-Gutierrez, S., Cordero, O. X., McFall-Ngai, M., Polz, M. F., et al. (2016). A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J. 10, 2907–2917. doi: 10.1038/ismej.2016.69
Bright, M., and Bulgheresi, S. (2010). A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8, 218–230. doi: 10.1038/nrmicro2262
Brown, M. V., Lauro, F. M., DeMaere, M. Z., Muir, L., Wilkins, D., Thomas, T., et al. (2012). Global biogeography of SAR11 marine bacteria. Mol. Syst. Biol. 8:595. doi: 10.1038/msb.2012.28
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 6, 80–92. doi: 10.4161/fly.19695
Darling, A. E., Jospin, G., Lowe, E., Matsen, F. A., Bik, H. M., and Eisen, J. A. (2014). PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2:e243. doi: 10.7717/peerj.243
Davis, M. P., Sparks, J. S., and Smith, W. L. (2016). Repeated and widespread evolution of bioluminescence in marine fishes. PLoS ONE 11:0155154. doi: 10.1371/journal.pone.0155154
Dunlap, P. V., Ast, J. C., Kimura, S., Fukui, A., Yoshino, T., and Endo, H. (2007). Phylogenetic analysis of host-symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23, 507–532. doi: 10.1111/j.1096-0031.2007.00157.x
Dunlap, P. V., Gould, A. L., Wittenrich, M. L., and Nakamura, M. (2012). Symbiosis initiation in the bacterially luminous sea urchin cardinalfish Siphamia versicolor. J. Fish Biol. 81, 1340–1356. doi: 10.1111/j.1095-8649.2012.03415.x
Dunlap, P. V., and Nakamura, M. (2011). Functional morphology of the luminescence system of Siphamia versicolor (Perciformes: Apogonidae), a bacterially luminous coral reef fish. J. Morphol. 272, 897–909. doi: 10.1002/jmor.10956
Dunlap, P. V., and Urbanczyk, H. (2013). “Luminous bacteria BT - the prokaryotes: prokaryotic physiology and biochemistry,” in The Prokaryotes: Prokaryotic Physiology and Biochemistry, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer Berlin Heidelberg).
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Gon, O., and Allen, G. R. (2012). Revision of the Indo-Pacific cardinalfish genus Siphamia (Perciformes: Apogonidae). Zootaxa 3294, 1–84. doi: 10.11646/zootaxa.3294.1.1
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gould, A. L., Dougan, K. E., Koenigbauer, S. T., and Dunlap, P. V. (2016). Life history of the symbiotically luminous cardinalfish Siphamia tubifer (Perciformes: Apogonidae). J. Fish Biol. 89, 1359–1377.
Gould, A. L., and Dunlap, P. V. (2019). Shedding light on specificity: population genomic structure of a symbiosis between a coral reef fish and luminous bacterium. Front. Microbiol. 10:2670. doi: 10.3389/fmicb.2019.02670
Guerrero-Ferreira, R. C., and Nishiguchi, M. K. (2007). Biodiversity among luminescent symbionts from squid of the genera Uroteuthis, Loliolus and Euprymna (Mollusca: Cephalopoda). Cladistics 23, 497–506. doi: 10.1111/j.1096-0031.2007.00155.x
Heard, S. B., and Hauser, D. L. (1995). Key evolutionary innovations and their ecological mechanisms. Hist. Biol. 10, 151–173. doi: 10.1080/10292389509380518
Hendry, T. A., de Wet, J. R., and Dunlap, P. V. (2014). Genomic signatures of obligate host dependence in the luminous bacterial symbiont of a vertebrate. Environ. Microbiol. 16, 2611–2622. doi: 10.1111/1462-2920.12302
Hendry, T. A., Freed, L. L., Fader, D., Fenolio, D., Sutton, T. T., and Lopez, J. V. (2018). Ongoing transposon-mediated genome reduction in the luminous bacterial symbionts of deep-sea ceratioid anglerfishes. MBio 9:e01017–17. doi: 10.1128/mBio.01033-18
Herrera, M., Klein, S. G., Campana, S., Chen, J. E., Prasanna, A., Duarte, C. M., et al. (2020). Temperature transcends partner specificity in the symbiosis establishment of a cnidarian. ISME J. 1013:1–13. doi: 10.1038/s41396-020-00768-y
Hykin, S. M., Bi, K., and McGuire, J. A. (2015). Fixing formalin: a method to recover genomic-scale DNA sequence data from formalin-fixed museum specimens using high-throughput sequencing. PLoS ONE 10:e0141579. doi: 10.1371/journal.pone.0141579
Jain, C., Rodriguez, -R L. M, Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jombart, T., Balloux, F., and Dray, S. (2010). adephylo: exploratory analyses for the phylogenetic comparative method. Bioinformatics 26, 1–21. doi: 10.1093/bioinformatics/btq292
Jones, B. W., Lopez, J. E., Huttenburg, J., and Nishiguchi, M. K. (2006). Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Mol. Ecol. 15, 4317–4329. doi: 10.1111/j.1365-294X.2006.03073.x
Joy, J. B. (2013). Symbiosis catalyses niche expansion and diversification. Proc. R. Soc. B Biol. Sci. 280:20122820. doi: 10.1098/rspb.2012.2820
Kaeding, A. J., Ast, J. C., Pearce, M. M., Urbanczyk, H., Kimura, S., Endo, H., et al. (2007). Phylogenetic diversity and cosymbiosis in the bioluminescent symbioses of “Photobacterium mandapamensis”. Appl. Environ. Microbiol. 73, 3173–3182. doi: 10.1128/AEM.02212-06
Koch, E. J., Miyashiro, T., McFall-Ngai, M. J., and Ruby, E. G. (2014). Features governing symbiont persistence in the squid–vibrio association. Mol. Ecol. 23, 1624–1634. doi: 10.1111/mec.12474
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Lee, K. H., and Ruby, E. G. (1994). Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60, 1565–1571. doi: 10.1128/AEM.60.5.1565-1571.1994
Li, H. (2013). Aligning Sequence Reads, Clone Sequences and Assembly Contigs With BWA-MEM. arXiv preprint arXiv:1303.3997 http://arxiv.org/abs/1303.3997
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). 1000 genome project data., Abecasis G., Durbin R.; 1000 Genome Project Data 000 Genome Project Data 000 Genome Project Data Processing Subgroup. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Matias, A. M., and Hereward, J. (2018). The complete mitochondrial genome of the five-lined cardinalfish Cheilodipterus quinquelineatus (Apogonidae). Mitochondrial DNA Part B, 3, 521–522. doi: 10.1080/23802359.2018.1467221
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol., 32, 268–274. doi: 10.1093/molbev/msu300
Nishiguchi, M. K. (2000). Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl. Environ. Microbiol. 66, 3550–3555. doi: 10.1128/AEM.66.8.3550-3555.2000
Nishiguchi, M. K., and Nair, V. S. (2003). Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. Int. J. Syst. Evol. Microbiol. 53, 2019–2026. doi: 10.1099/ijs.0.02792-0
Norris, R. D. (1996). Symbiosis as an evolutionary innovation in the radiation of Paleocene planktic foraminifera. Paleobiology 22, 461–480. doi: 10.1017/S0094837300016468
Norsworthy, A. N., and Visick, K. L. (2013). Gimme shelter: how Vibrio fischeri successfully navigates an animal's multiple environments. Front. Microbiol. 4, 356. doi: 10.3389/fmicb.2013.00356
Nyholm, S. V., and McFall-Ngai, M. (2004). The winnowing: establishing the squid–Vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. doi: 10.1038/nrmicro957
Padgham, M., and Sumner, M. D. (2020). geodist: Fast, Dependency-Free Geodesic Distance Calculations. R package version 0.0.4. Available online at: https://CRAN.R-project.org/package=geodist
Portik, D. M., and Wiens, J. J. (2020). SuperCRUNCH: A bioinformatics toolkit for creating and manipulating supermatrices and other large phylogenetic datasets. Methods Ecol. Evol. 11, 763–772. doi: 10.1111/2041-210X.13392
Reichelt, J. L., Nealson, K., and Hastings, J. W. (1977). The specificity of symbiosis: pony fish and luminescent bacteria. Arch. Microbiol. 112, 157–161. doi: 10.1007/BF00429329
Revell, L. (2011). Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Ronquist, F. (1998). Phylogenetic approaches in coevolution and biogeography. Zool. Scr. 26, 313–322. doi: 10.1111/j.1463-6409.1997.tb00421.x
Rotman, E. R., Bultman, K. M., Brooks, J. F., Gyllborg, M. C., Burgos, H. L., Wollenberg, M. S., et al. (2019). Natural strain variation reveals diverse biofilm regulation in squid-colonizing Vibrio fischeri. J. Bacteriol. 201:e00033-19. doi: 10.1128/JB.00033-19
Ruane, S., and Austin, C. C. (2017). Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Mol. Ecol. Resources, 17, 1003–1008. doi: 10.1111/1755-0998.12655
Seemann, T. (2015). Snippy: Fast Bacterial Variant Calling From NGS Reads. Snippy: fast bacterial variant calling from NGS reads.
Sul, W. J., Oliver, T. A., Ducklow, H. W., Amaral-Zettler, L. A., and Sogin, M. L. (2013). Marine bacteria exhibit a bipolar distribution. Proc. Natl. Acad. Sci. U.S.A. 110, 2342–2347. doi: 10.1073/pnas.1212424110
Thacker, C. E., and Roje, D. M. (2009). Phylogeny of cardinalfishes (Teleostei: Gobiiformes: Apogonidae) and the evolution of visceral bioluminescence. Mol. Phylogenet. Evol. 52, 735–745. doi: 10.1016/j.ympev.2009.05.017
Troussellier, M., Escalas, A., Bouvier, T., and Mouillot, D. (2017). Sustaining rare marine microorganisms: macroorganisms as repositories and dispersal agents of microbial diversity. Front. Microbiol. 8:947. doi: 10.3389/fmicb.2017.00947
Tyberghein, L., Verbruggen, H., Pauly, K., Troupin, C., Mineur, F., and De Clerck, O. (2012). Bio-ORACLE: a global environmental dataset for marine species distribution modelling. Global Ecol. Biogeogr. 21, 272–281. doi: 10.1111/j.1466-8238.2011.00656.x
Urbanczyk, H., Ogura, Y., Hendry, T. A., Gould, A. L., Kiwaki, N., Atkinson, J. T., et al. (2011). Genome sequence of Photobacterium mandapamensis strain svers. 1.1, the bioluminescent symbiont of the cardinal fish Siphamia versicolor. J. Bacteriol. 193, 3144–3145. doi: 10.1128/JB.00370-11
Wada, M., Kamiya, A., Uchiyama, N., Yoshizawa, S., Kita-Tsukamoto, K., Ikejima, K., et al. (2006). LuxA gene of light organ symbionts of the bioluminescent fish Acropoma japonicum (Acropomatidae) and Siphamia versicolor (Apogonidae) forms a lineage closely related to that of Photobacterium leiognathi ssp. mandapamensis. FEMS Microbiol. Lett. 260, 186–192. doi: 10.1111/j.1574-6968.2006.00322.x
Wägele, H. (2004). Potential key characters in Opisthobranchia (Gastropoda, Mollusca) enhancing adaptive radiation. Org. Divers. Evol. 4, 175–188. doi: 10.1016/j.ode.2004.03.002
Wallin, I. E. (1927). Symbionticism and the Origin of Species. Baltimore: Williams and Wilkins company.
Wolfe, A. J., Millikan, D. S., Campbell, J. M., and Visick, K. L. (2004). Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70, 2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004
Wollenberg, M. S., and Ruby, E. G. (2009). Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl. Environ. Microbiol. 75, 193–202. doi: 10.1128/AEM.01792-08
Keywords: symbiosis, bioluminescence, coral reef fish, photobacterium, Siphamia tubifer, symbiont specificity, co-diversification, museum genomics
Citation: Gould AL, Fritts-Penniman A and Gaisiner A (2021) Museum Genomics Illuminate the High Specificity of a Bioluminescent Symbiosis for a Genus of Reef Fish. Front. Ecol. Evol. 9:630207. doi: 10.3389/fevo.2021.630207
Received: 17 November 2020; Accepted: 08 January 2021;
Published: 04 February 2021.
Edited by:
Adrienne M. S. Correa, Rice University, United StatesReviewed by:
Tory Arminta Hendry, Cornell University, United StatesLydia Jeanne Baker, Oregon State University, United States
Copyright © 2021 Gould, Fritts-Penniman and Gaisiner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alison L. Gould, agould@calacademy.org
 Alison L. Gould
Alison L. Gould Allison Fritts-Penniman
Allison Fritts-Penniman Ana Gaisiner
Ana Gaisiner