How the Devil Ray Got Its Horns: The Evolution and Development of Cephalic Lobes in Myliobatid Stingrays (Batoidea: Myliobatidae)
- 1Department of Biology, San Francisco State University, San Francisco, CA, United Statess
- 2Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, United States
- 3Virginia Institute of Marine Science, College of William and Mary, Williamsburg, VA, United States
Manta rays and their relatives of the family Myliobatidae have pectoral fins that have been modified for underwater flight, as well as a pair of fleshy projections at the anterior of the body called cephalic lobes, which are specialized for feeding. As a unique trait with a dedicated function, cephalic lobes offer an excellent opportunity to elucidate the processes by which diverse body plans and features evolve. To shed light on the morphological development and genetic underpinnings of cephalic lobes, we examined paired fin development in cownose rays, which represent the sister taxon to manta rays in the genus Mobula. We find that cephalic lobes develop as anterior pectoral fin domains and lack independent posterior patterning by 5′ HoxD genes and Shh, indicating that cephalic lobes are not independent appendages but rather are modified pectoral fin domains. In addition, by leveraging interspecies comparative transcriptomics and domain-specific RNA-sequencing, we identify shared expression of anterior patterning genes, including Alx1, Alx4, Pax9, Hoxa13, Hoxa2, and Hoxd4, in the pectoral fins of cownose ray (Rhinoptera bonasus) and little skate (Leucoraja erinacea), providing evidence supporting homology between the cephalic lobes of myliobatids and the anterior pectoral fins of skates. We also suggest candidate genes that may be involved in development of myliobatid-specific features, including Omd, which is likely associated with development of thick anterior pectoral fin radials of myliobatids, and Dkk1, which may inhibit tissue outgrowth at the posterior boundary of the developing cephalic lobes. Finally, we observe that cephalic lobes share a surprising number of developmental similarities with another paired fin modification: the claspers of male cartilaginous fishes, including enrichment of Hand2, Hoxa13, and androgen receptor. These results suggest that cephalic lobes may have evolved by co-opting developmental pathways that specify novel domains in paired fins. Taken together, these data on morphological development and comparative gene expression patterns illustrate how distinct body plans and seemingly novel features can arise via subtle changes to existing developmental pathways.
Introduction
Manta rays (also known as devil rays) and their relatives of the family Myliobatidae exhibit modified body plan features that arose in association with the evolution of oscillatory swimming and invasion of the pelagic environment (Rosenberger, 2001; Schaefer and Summers, 2005; Mulvany and Motta, 2013; Hall et al., 2018). While myliobatids share classic components of the batoid body plan, including dorsoventral compression and pectoral fins that expand anteriorly and fuse to the head, they also exhibit derived morphological modifications that are unique to the family, such as anterior projections called “cephalic lobes” that are used to locate and manipulate prey during feeding (Sasko et al., 2006; Mulvany and Motta, 2014), loss of anterior-posterior disc symmetry typical of most skates and rays (Hall et al., 2018) and laterally extended pectoral fins that arose in association with oscillatory swimming (Fontanella et al., 2013; Franklin et al., 2014; Martinez et al., 2016). While most batoids use their broad disc or diamond-shaped pectoral fins for both swimming and feeding, the myliobatids exhibit functional separation of swimming and feeding behaviors into the pectoral fins and cephalic lobes, respectively, which have been optimized for those specific tasks (Mulvany and Motta, 2013; Hall et al., 2018).
The separation of structures dedicated to feeding and locomotion appears to be associated with a large-scale remodeling of the myliobatid body plan, particularly in the pectoral fins. For example, the skeletal elements in the anterior pectoral fins of myliobatids fuse at the interradial joints to form the compagibus laminam, a plate of skeletal elements that likely promotes lift during steady swimming (Schaefer and Summers, 2005; Hall et al., 2018). In contrast, fin rays in the posterior pectoral fins of myliobatids are thinner and more numerous than those of non-myliobatids, which likely contributes to increased maneuverability (Hall et al., 2018). In addition, myliobatid pectoral fins are laterally expanded, resulting in a high aspect ratio that reduces drag while increasing lift and thrust generation (Fontanella et al., 2013; Franklin et al., 2014), the center of mass is shifted anteriorly, which stabilizes the body during steady swimming (Fontanella et al., 2013), and the pectoral fins are stiffened by calcification patterns in the radials that redistribute load-bearing elements and increase the power of high amplitude pectoral strokes (Schaefer and Summers, 2005). Together, these modifications are associated with the evolution of oscillatory swimming, or underwater flight, the primary mode of locomotion in myliobatids.
Though these morphological adaptations promote efficient long-range swimming in the pelagic environment, they also likely represent constraints on feeding, as stiffened pectoral fins are not well-suited for the prey capture strategies observed in other batoids, such as tenting prey against the substrate (Wilga et al., 2012; Mulvany and Motta, 2014), which requires flexibility at the anterior of the pectoral fins. As such, myliobatids have evolved cephalic lobes, fleshy anterior appendages that are used primarily for feeding and exhibit a degree of flexibility that is comparable to the anterior pectoral fins of other batoids (Mulvany and Motta, 2013).
Cephalic lobes (sometimes referred to as cephalic fins) exhibit a variety of shapes, sizes, and functions (Nishida, 1990; Miyake et al., 1992; Mulvany and Motta, 2013). The most widely recognized example of cephalic lobes occurs in manta rays of the genus Mobula (Aschliman, 2014; Poortvliet et al., 2015; White et al., 2017), in which the elongated cephalic lobes unfurl to funnel plankton into the mouth when feeding (Notarbartolo-Di-Sciara, 1987; Notarbartolo-di-Sciara and Hillyer, 1989; Ari and Correia, 2008). In cownose rays (Rhinoptera), the flexible paired cephalic lobes are fused at the midline and used to trap prey against the substrate, analogous to the tenting behavior performed by the anterior pectoral fins of other batoids (Sasko et al., 2006; Fisher et al., 2011; Mulvany and Motta, 2014), and in bat rays (Myliobatis) and eagle rays (Aetobatus and Aetomylaeus), the cephalic lobes are fused into a single structure that is used like a shovel to unearth benthic prey (Mulvany and Motta, 2013). Cephalic lobes are covered in electrosensory pores that are used for locating hidden prey in all myliobatid genera except Mobula, which lack pores (Jordan, 2008; Mulvany and Motta, 2013; Bedore et al., 2014). By adopting key functions associated with feeding, such as prey detection and capture, cephalic lobes may have released the pectoral fins from constraints associated with feeding, thereby facilitating the evolution of oscillatory swimming in the Myliobatidae and subsequent invasion of the pelagic environment.
Despite their significant role in myliobatid ecology and evolution, the mechanisms underlying the evolution and development of cephalic lobes remain a mystery. Myliobatid taxa with paired cephalic lobes have been referred to as the only vertebrates with three pairs of functional appendages (Nelson et al., 2016). If cephalic lobes are independent appendages, then we would expect them to develop separately from the pectoral fins and to exhibit independent canonical expression of genes that pattern paired appendages, such as posterior patterning by 5′ HoxD genes (see Kmita et al., 2005; Freitas et al., 2007, 2012) and Shh expression (see Krauss et al., 1993; Riddle et al., 1993; Chang et al., 1994; Dahn et al., 2007). However, most examples of novel morphological traits arise from existing or duplicated structures that are modified to the extent that homology becomes obscured. As such, cephalic lobes may have evolved as modifications to the anterior pectoral fins, in which case we would expect them to develop in association with anterior pectoral fins, lack independent posterior canonical fin/limb patterning and to share homologous gene expression patterns with the anterior pectoral fins of other batoids that lack cephalic lobes. Cephalic lobes, therefore, represent an excellent opportunity to inform the mechanisms by which unique morphological features evolve in association with novel functions and ecological roles.
Because manta rays (genus Mobula) are exceptionally challenging to sample, and many mobulids are listed as vulnerable or endangered (Couturier et al., 2012), we focused our efforts on a representative of their sister taxon: cownose rays of the genus Rhinoptera, which exhibit a similar body plan, including paired cephalic lobes. To elucidate the morphological and molecular evolution and development of myliobatid cephalic lobes, we sampled embryos of the cownose ray, Rhinoptera bonasus, during outgrowth and patterning of the pectoral fins and characterized their developmental progression. To evaluate homology between myliobatid cephalic lobes and the anterior pectoral fins of a batoid that lacks cephalic lobes, we sequenced RNA from developing cownose ray cephalic lobes and pectoral fins for comparison with gene expression patterns in developing pectoral fins of the little skate (Nakamura et al., 2015) using similar methodology. Using this dual-pronged approach, we addressed the following specific questions:
1) Do cephalic lobes develop as independent paired appendages or do they develop as modified anterior pectoral fins?
2) Are myliobatid cephalic lobes and rajid anterior pectoral fins patterned by shared developmental gene expression that would support homology?
3) Can we identify distinct developmental processes and gene expression patterns associated with myliobatid-specific features, including cephalic lobes?
To our knowledge, this is the first study to document fin development in the myliobatid lineage. Our results indicate that cephalic lobes do not develop as independent appendages, but rather as modifications of the anterior pectoral fins which share homology with the anterior pectoral fins of skates. We also offer a description of the processes underlying development of myliobatid pectoral fins and cephalic lobes, which includes distinct phases of anterior expansion followed by fusion to the pharyngeal basket, head, and rostrum. We further identify gene expression patterns likely associated with myliobatid-specific pectoral fin modifications, including specification of cephalic lobes, the compagibus laminam, and a myliobatid-specific developmental feature we call the “notch.” Finally, we found enrichment of shared genes during development of both claspers and cephalic lobes which also share several morphological features, suggesting a redeployment of pathways that specify development of these paired fin modifications.
Methods
Sample Collection and Developmental Staging
It is intractable to sample myliobatid embryos at specific developmental stages, as there is no published developmental timeline for any myliobatid taxon and no appropriately comparable model taxa have been characterized. Therefore, we conducted a pilot study to delineate the window of pectoral fin development in cownose rays from a wild population in Chesapeake Bay. Then, working with the bycatch of commercial fishermen, we sampled during July and August of 2016 and 2017. Embryos used for RNA-sequencing were preserved in RNALater, incubated at 4°C for 24–72 h, and then stored at −20°C until dissection and RNA extraction. Samples used for staging were fixed in 4% paraformaldehyde for 48 h before being transferred to methanol and stored at −80°C. Embryos were staged in an RNase-free environment, grouped by characters that we observed to be consistently associated with development of the pectoral fins (Table 1), and assigned a numerical index representing these characters (Table 2).
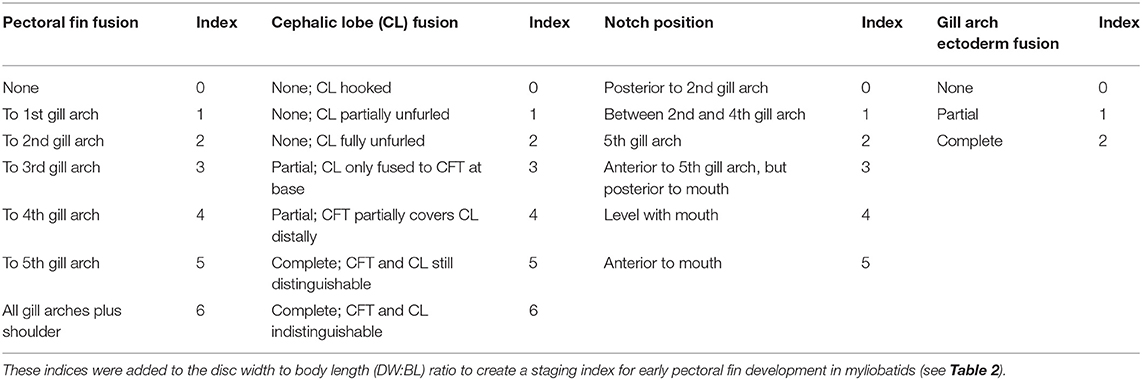
Table 1. Developmental index parameters based on characters associated with development of cownose ray pectoral fins.
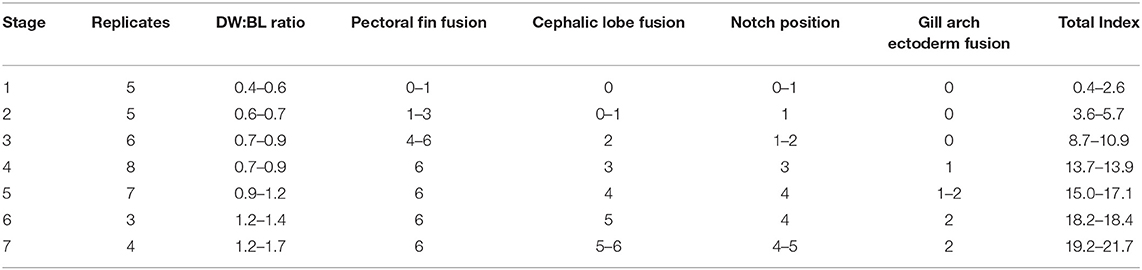
Table 2. Characters evaluated in developmental index and range of scores for each stage (See Table 1).
One goal of this study was to evaluate homology between the anterior pectoral fins of skates and the cephalic lobes of myliobatids using comparative transcriptomics. Because developmental-genetic programs can shift in space and time, it is important to examine similar developmental stages in each taxon. Therefore, morphological characters that were shared between developing cownose ray and winter skate (Leucoraja ocellata, following Maxwell et al., 2008) pectoral fins were identified. These were used to determine the most comparable stages of development in the cownose ray, Rhinoptera bonasus (R. bonasus), and little skate, Leucoraja erinacea (L. erinacea) (Table S1) for gene expression analysis.
Of all stages collected, the stage three R. bonasus embryos we sampled for RNA-sequencing represented the most biological replicates (six), and therefore gave us the greatest power to detect differentially expressed genes (DEGs). In addition, the pectoral fins undergo significant developmental processes during this stage, including anterior expansion, similar to stage 31 L. erinacea embryos sequenced by Nakamura et al. (2015). Due to the enhanced power to detect DEGs, the active developmental processes (including anterior expansion of the pectoral fins), and the availability of comparable data in L. erinacea, we focused our molecular analyses on stage three of R. bonasus development.
RNA Sequencing of Distinct Pectoral Fin Domains
Nakamura et al. (2015) sequenced RNA from three equal-sized pectoral fin domains of developing embryos of L. erinacea, a representative batoid from the skate family (Rajidae), which diverged from all other batoids approximately 180 million years ago (Aschliman et al., 2012). We used these data for comparison to address the question of cephalic lobe/pectoral fin homology. To replicate the methods of Nakamura et al. (2015), we cut the left pectoral fin of stage three R. bonasus embryos (which includes the cephalic lobe) in thirds. We then added a layer of resolution to our dissections by bisecting each third of the pectoral fin again. The cephalic lobe comprised a distinct domain when the fin was split in this way (Figure 1). Further, when we included RNA-sequencing reads from adjacent dissections, our data represented three evenly-sized pectoral fin domains (following Nakamura et al., 2015; see Figure 1) with which to evaluate homology with the three domains previously examined in L. erinacea.
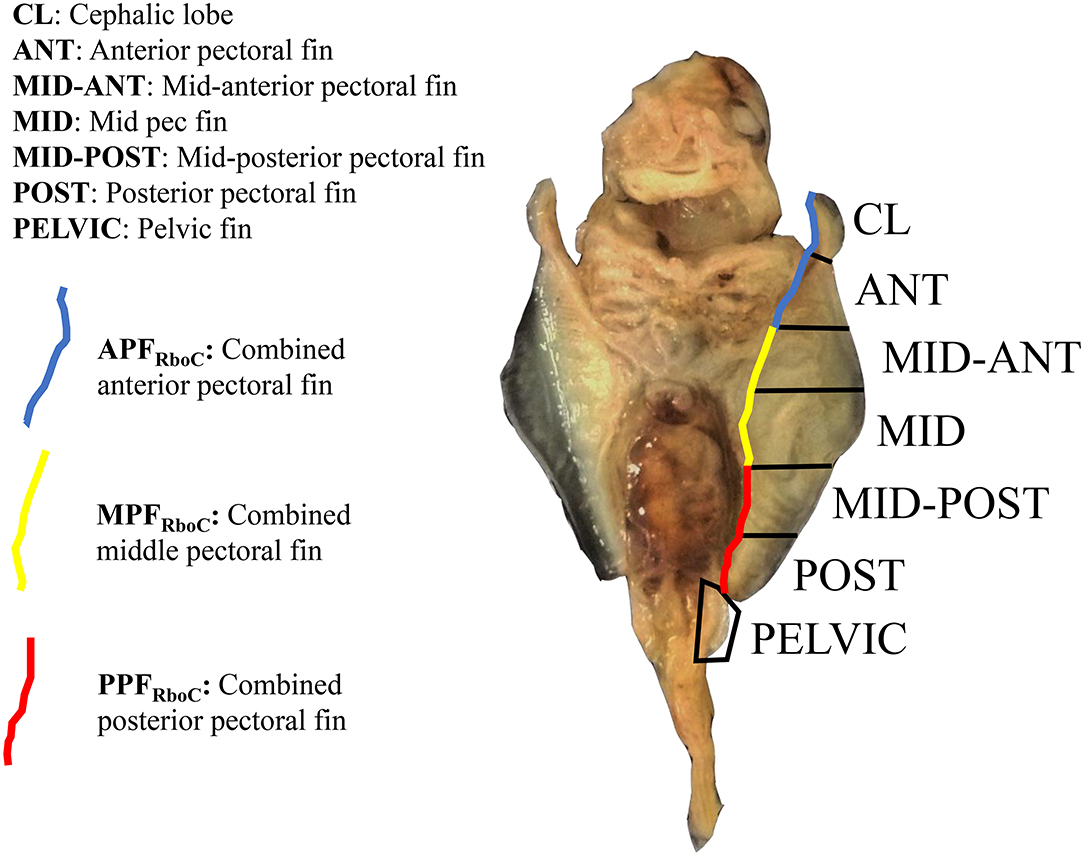
Figure 1. Extraction domains for RNA-sequencing of stage three cownose ray embryos. View is ventral; anterior is at top. The left pectoral fin was cut into six evenly-sized pieces and separate libraries with distinct barcodes were prepared for each fin domain. Later, differential expression scores were averaged from adjacent domains for interspecies comparisons with the little skate (blue, yellow, red). RNA was also extracted from the left pelvic fin and from the area actively undergoing fusion between the pectoral fin and gill arches (not shown).
Following dissection, RNA was extracted from the six R. bonasus pectoral fin domains, as well as two additional fin domains, including pelvic fin. We extracted RNA using Trizol and followed with column-based purification using the PureLink RNA mini kit (Invitrogen) and a subsequent DNase I digestion. Purity was assessed using a NanoDrop with 260/280 ratios registering between 2.0 and 2.5. When there was residual guanidine salt, as indicated by a 260/230 ratio < 1.5, a SPRI bead purification was conducted and the eluate was reanalyzed to confirm that the final 260/230 ratio was greater than 1.5.
Sample aliquots were diluted and RNA integrity numbers (RIN) were evaluated using an RNA Pico chip on an Agilent BioAnalyzer. All samples exhibited RIN scores >8. Total RNA from each sample was quantified using a Qubit and 1 μg of RNA from each sample was used for Poly(A) mRNA enrichment using NEBNext oligo d(T) magnetic beads. After mRNA isolation and fragmentation, cDNA libraries were constructed using the NEBNext Ultra Directional Library Prep Kit for Illumina and each sample was given a unique barcode. Double-sided size selection was performed using SPRI beads targeting 400 bp fragments. Fragment size, concentration, and library purity were verified using a High Sensitivity DNA chip on a BioAnalyzer. In total, 48 libraries representing eight distinct fin domains with six biological replicates were normalized, pooled, and sequenced on an Illumina HiSeq 4000 using 100 bp paired end (PE) reads. Final library quality assessment and sequencing were conducted at the University of California, Berkeley; all other labwork occurred at San Francisco State University.
The cownose ray is a non-model organism, and the closest taxon for which a published genome exists is the elephant shark (Venkatesh et al., 2014), which diverged from the cownose ray and other elasmobranchs about 420 million years ago (Inoue et al., 2010). When sequence divergence is >15%, de novo transcriptome assemblies are recommended over reference-based mapping approaches (Vijay et al., 2013). To aid the de novo assembly process, we sequenced additional cephalic lobe, clasper, and pectoral fin domains on an Illumina MiSeq with longer reads (250 and 300 bp PE). The longer reads facilitated de novo transcriptome assembly by helping to bridge low complexity and redundant regions to which the shorter HiSeq reads could map. These samples were not included in our DGE analyses because they were prepared and sequenced using distinct kits and methodologies, which may introduce significant biases in expression levels (van Dijk et al., 2014).
Data Analysis
Data Preprocessing
Adapters and low quality sequences from cownose ray data were trimmed from fastq files using the TrimGalore (Krueger, 2012) wrapper for Cutadapt (Martin, 2011) and FastQC (Andrews, 2010). Overlapping reads were combined with Flash (Magoc and Salzberg, 2011) prior to assembly and orphaned reads were retained. To reduce the impact of genetic variability, data from fin domains of only three individuals were used in the assembly, including: two regions of a stage one embryo (broad anterior pectoral fin region and mid-anterior pectoral fin), one region of a stage seven embryo (clasper) and eight regions of stage three embryos (cephalic lobe, anterior pectoral fin, mid-anterior pectoral fin, mid-pectoral fin, mid-posterior pectoral fin, posterior pectoral fin, zipper region, and pelvic fin). Read counts before and after filtering are reported in Table S2.
Transcriptome Assembly and Annotation
Sequencing with both HiSeq (stage 3 samples, 100 bp PE reads) and MiSeq (stage 1 samples, stage 7 sample, 300 bp PE reads) was conducted to help generate full length transcripts. Abyss (Simpson et al., 2009) was used to assemble contiguous sequences (contigs) from the paired end (PE) and orphaned sequence data with 10 km sizes ranging from 25 to 70 nucleotides. Contigs of 500 nucleotides or greater were summarized according to similarity (e-value < 1e-160) with BLAST+ tools (Camacho et al., 2009) into 63,441 unique contigs.
Using iterative BLAST searches and pairwise alignments, contig sequences with strong e-values (<1e-10) were annotated and used in the final transcriptome. Specifically, initial screening and annotation of contigs was conducted iteratively by best-reciprocal-blast using peptide, coding sequence, and non-coding libraries from Danio rerio (GRCz10) with BLAST+ tools and a minimum e-value threshold of 0.1. Each R. bonasus contig was paired with D. rerio sequences and the lowest average e-value was chosen as the annotation. Therefore, more than one R. bonasus contig could be designated with the same D. rerio sequence, allowing for potential redundancy in the de novo R. bonasus transcriptome assembly. The longest open reading frames (ORFs) that were unannotated to D. rerio sequences were annotated by BLASTp using the RefSeq vertebrate protein database (downloaded 4-4-2017). Remaining unannotated contigs were designated by their top forward BLAST hit in D. rerio, resulting in a total transcriptome size of 27,900 contigs. For consistency, the same process was used in generating the L. erinacea transcriptome from public stage 31 embryo L. erinacea 100 bp PE RNA-seq data (SRA, PRJNA288370, Nakamura et al., 2015), generating 24,006 transcripts.
Annotations of genes highlighted in Figures 4 and 5 were individually verified using NCBI's BLASTn (Altschul et al., 1990). We examined the top vertebrate hits, including, but not limited to, the elephant shark (Callorhincus milli), zebrafish (Danio rerio), coelacanth (Latimeria chalumnae), and mouse (Mus musculus), in addition to the catshark (Scyliorhinus sp.) when sequence data were available. BLAST e-values and accession numbers for the sequences to which the R. bonasus and L. erinacea transcripts most reliably aligned are reported in Table S3. Transcripts that were annotated as a different gene in more than one taxon were excluded from analysis due to potential ambiguity in the annotation. One exception was Ntrk2, which had the highest overall BLAST score when aligned with Bdnf from the whale shark, Rhinocodon typus (e-value = 0.00; Accession XM_020520327.1), but otherwise preferentially aligned with Ntrk2 from the other taxa evaluated (Table S3). However, the two genes function in tandem: Ntrk2 is a receptor that shows a high affinity for binding Bdnf (Agerman, 2003) and has been used as an indicator of Bdnf activity (Pita-Thomas et al., 2010), so we did not discard this gene. Due to their highly conserved homeodomain (Krumlauf, 1994), we were able to translate and align all of the various Hox genes highlighted in the text and phylogenetically confirm the correct annotation of Hox genes using Geneious 11.1.15 (Figure S1).
Alignment and Additional Filtering
R base (Ihaka and Gentleman, 1996) and Bioconductor (Gentleman et al., 2004) software packages were used for alignment of quality filtered PE data and differential expression comparisons. Strand-specific alignment was conducted using the seed-and-vote methodology for uniquely aligned reads, as implemented in the Rsubread package (Liao et al., 2013), to the annotated R. bonasus transcriptome. Rsamtools was used for file indexing and manipulation (Morgan et al., 2018). Due to potential biases unrelated to developmental gene expression differences, contig sequences annotated as ribosomal, mitochondrial, predicted, hypothetical, location, or uncharacterized were removed from consideration in differential expression. Moreover, only contigs that were represented by more than 100 reads across the full data set of 48 samples and more than 50 reads in at least one individual were considered reasonable to use for differential expression comparisons such that 11,881 transcripts were considered in the differential expression comparisons. Data from L. erinacea and R. bonasus were processed similarly, except that in L. erinacea, the filter required at least 50 reads across all samples (after removing highest) and at least one sample that had at least 25 reads, which was used to help minimize the effect of outliers in the smaller sample set (N = 8). In total, 12,822 L. erinacea transcripts were considered in the differential expression comparisons.
Differential Expression
Normalization of read count data for the filtered transcripts was conducted with the trimmed mean of M-values (TMM) method (Robinson and Oshlack, 2010). EdgeR (Robinson et al., 2010) was used to conduct quasi-likelihood F-tests between groups of samples and false discovery rate adjusted p-values (Benjamini-Hochberg) were used to control for multiple testing.
Interspecies Comparisons
For interspecies comparisons, we included RNA-sequencing reads from adjacent domains in R. bonasus (Figure 1) to replicate the three evenly-sized pectoral fin domains sequenced by Nakamura et al. (2015) in L. erinacea. We created lists of differentially expressed genes in each species separately and queried the lists of DEGs for genes known to be associated with paired fin development in L. erinacea and other non-batoid taxa. This approach accounts for bias associated with library preparation, sequencing, and counting efficiency by comparing ratios of differentially expressed genes among fin domains within each species separately (according to Dunn et al., 2013).
Results
Patterns of Development in Cownose Ray Pectoral Fins
The stages of cownose ray fin development documented here are roughly analogous to stages 28–31 in bamboo sharks and catsharks (Ballard et al., 1993; Onimaru et al., 2018), though we note that sharks lack the anterior expansion of the pectoral fins characteristic of the Batoidea. This anterior expansion appears to occur via similar mechanisms across batoid clades. In Raja eglanteria (Luer et al., 2007), Leucoraja ocellata (Maxwell et al., 2008), Urobatis halleri (Babel, 1966), and Rhinoptera bonasus (this study), pectoral fins begin developing as outgrowths from the body wall, as in all vertebrates. In skates and U. halleri, the anterior pectoral fins adopt a hook-like morphology and continue developing by growing away from the body (anterodistally) as they fuse to the pharyngeal basket in a rostral progression. The process terminates when the anterior pectoral fins fuse to the rostrum anterior to the mouth. Similarly, the anterior pectoral fins of the cownose ray begin developing posterior to the gill arches and assume a hook-like morphology. In the cownose ray, these hooks correspond to the developing cephalic lobes (Figure 2A). As development proceeds, the cephalic lobes unfurl but remain distinguished from the rest of the pectoral fins by a physical “notch,” where tissue outgrowth appears to be inhibited (Figure 2B). Concurrent with the unfurling of the cephalic lobes, the pectoral fins expand anterodistally and fuse to the body, beginning at the posterior pharyngeal arch and progressing rostrally (Figure 2C). Development to this point parallels the process observed in other batoids. However, once the region of the notch fuses at the shoulder near pharyngeal arch I, two processes commence in cownose ray pectoral fins that may be unique to myliobatids.
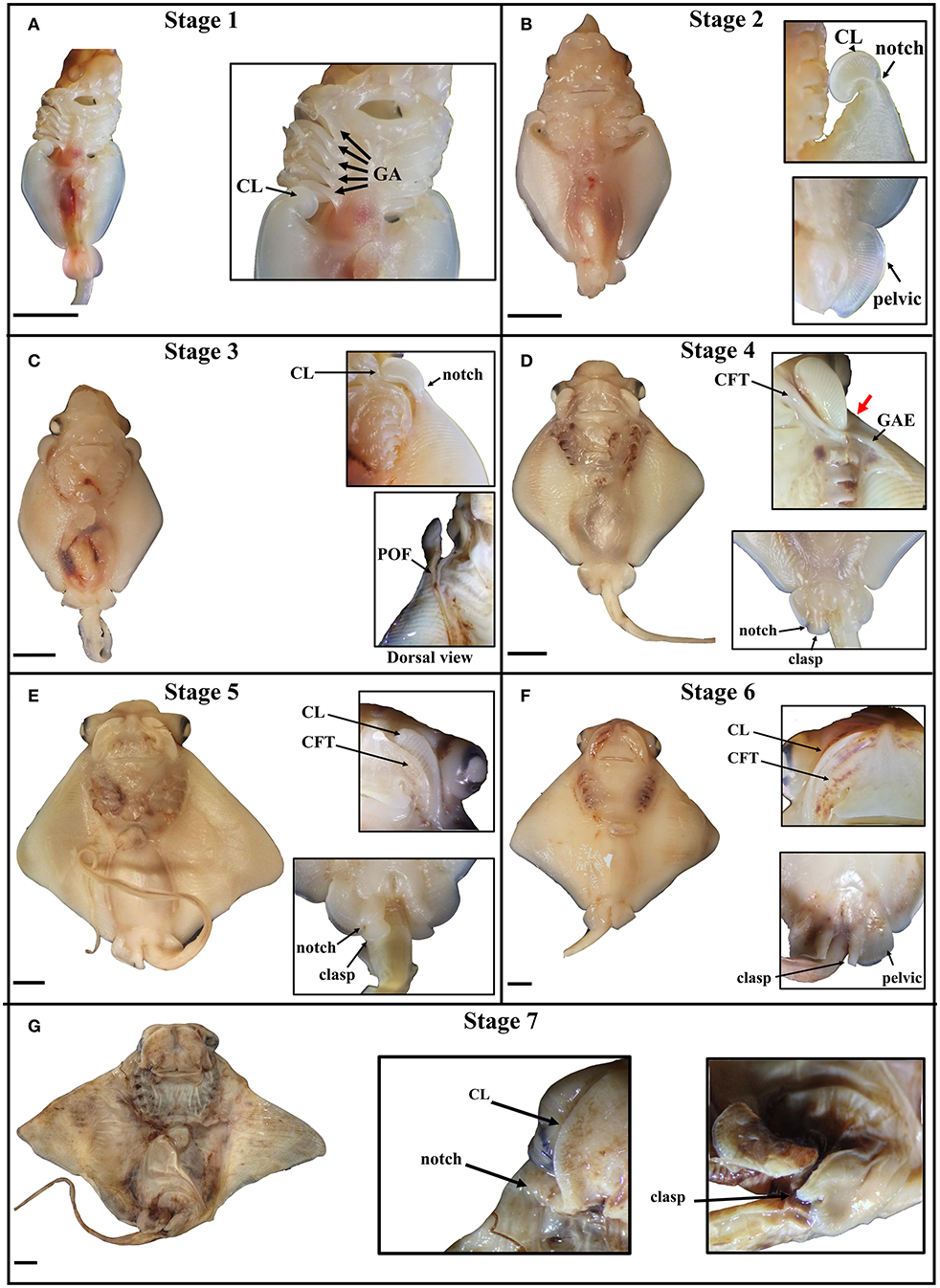
Figure 2. Stages of pectoral fin development in the cownose ray, Rhinoptera bonasus. Black bars are 5 mm. All pictures are of the ventral side unless indicated. Top of frame is anterior. Inset images do not necessarily come from the same whole specimen shown and may represent different individuals at the same developmental stage. CL, cephalic lobe; GA, gill arch; clasp, clasper; CFT, craniofacial tissue pocket. (A) Stage one: Cephalic lobes are curled posterior to the gill arches. Pectoral fins have only just begun to expand anteriorly. Pelvic fin is round, shaped like a tear drop. (B) Stage two: The cephalic lobes remain mostly curled, but have begun to unfurl from the pectoral fins, while a notch is becoming visibly distinct at the posterior boundary of each cephalic lobe. The pectoral fins expand anteriorly and distally, extending no further to the anterior than four gill arches, but they are not yet fused to the body. The pelvic fins are rounded half circles. (C) Stage three: The pectoral fins fuse to the body during this stage, with visible evidence of this process showing at the point of fusion (POF), or the zipper region. The cephalic lobes unfurl, aligning with the developing primary cartilage of the pectoral fins and assuming positions posterolaterally relative to the mouth. In late stage three embryos, the notch on each side begins to expand along the dorso-ventral axis, slightly rotating the cephalic lobes and shifting them ventrally. (D) Stage four: The pectoral fins are now fused to the body up to the notch, which deviates ventrally as it fuses (thick red arrow), positioning each cephalic lobe near the mouth. The unfused cephalic lobes settle into ventral pockets of craniofacial tissue (CFT) which themselves are connected to sheets of ectoderm that cover the gill arches. The gill arch ectodermal sheets (GAE) extend over the ventral pectoral fins at the anterior, assuming triangular shapes. In males, a notch forms in each pelvic fin, delineating the boundary between the pelvic fins and the developing claspers, which are shaped like two-dimensional paddles. (E) Stage five: The GAEs fuse to ventral ectodermal tissue on the pectoral fins, thereby completing the process of pectoral fin fusion posterior to the notch, though the cephalic lobes are not yet fully fused. The CFT pockets grow distally to incompletely cover the cephalic lobes, which are sandwiched between layers of dorsal and ventral CFT. Male claspers continue differentiating from the pelvic fins and begin to curl, appearing now as three-dimensional paddles, with proximal folding preceding distal. (F) Stage six: The CFT pockets cover the cephalic lobes, but are still distinguishable. The claspers begin to roll, appearing as distinct structures medial to the pelvic fins, which have assumed a squared morphology. (G) Stage seven: The cephalic lobes are now fully fused to the CFT pockets anterior to the mouth, completing the process of pectoral fin fusion. The claspers are fully rolled. The developing embryo now resembles a mature cownose ray and will continue to grow allometrically for another 9–10 months.
First, pockets of craniofacial tissue (CFT) begin developing lateral to the mouth (Figure 2D). These CFT pockets have ectodermal components that extend posteriorly to cover the gill arches, and distally to form a triangular sheet which eventually fuses to ectoderm of the pectoral fins. Second, the pattern of fusion near the notch begins to deviate ventrally to position the cephalic lobes within the CFT pockets on the ventral side of the body near the mouth. As development continues, the CFT pockets expand (Figure 2E), envelop (Figure 2F) and fuse to (Figure 2G) the cephalic lobes in a medial-distal progression that mirrors the caudal-rostral progression of pectoral fin fusion. By stage seven, the embryonic cownose ray has assumed the shape of an adult: fusion of pectoral fins, cephalic lobes, and CFT envelopment is complete, and only allometric growth remains. Surprisingly, the entire process of pectoral fin fusion spans just 2–3 weeks during the first month of the eleven-month cownose ray gestation period (J. Swenson, pers. obs.), indicating that the myliobatid body plan is established very early in development.
Cephalic Lobes Develop as Modified Anterior Pectoral Fins
In cownose rays, cephalic lobes do not develop from independent fin buds emanating from the head; rather, they develop as modifications to the anterior pectoral fins, closely resembling the hooks at the anterior of developing skate and ray pectoral fins (Babel, 1966; Luer et al., 2007; Maxwell et al., 2008). However, unlike the anterior pectoral fins of other batoids, cephalic lobes are distinguished from the rest of the pectoral fin by a small region of reduced tissue outgrowth we call the “notch.” Although most of the transient morphologies and processes observed during cephalic lobe development are also found in developing pectoral fins of other batoids (e.g., the anterior hook, fusion to the pharyngeal arches and rostrum), the notch has only been observed in cownose rays in association with cephalic lobe development, suggesting that the notch is a defining feature of cephalic lobe specification.
Another distinction between developing pectoral fins in other batoids relative to cownose rays is manifest in the pattern of tissue fusion. Whereas the anterior pectoral fins in other batoids fuse to the rostrum in the same plane as the pectoral fins, in cownose rays, the pattern of fusion involves a distinct ventral deviation while the pectoral fins are fusing to the shoulder region corresponding to the notch. Ultimately, the unfused cephalic lobes are positioned on the ventral side of the body lateral to the mouth and enveloped within pockets of craniofacial tissue. This process may be unique to myliobatids with ventrally-positioned cephalic lobes.
Shared Gene Expression Patterns in Batoid Paired Fin Development and Homology of Cephalic Lobes
After observing that myliobatid cephalic lobes develop as modified anterior pectoral fins, we used RNA-Sequencing (Wang et al., 2009) to compare gene expression profiles among pectoral fin domains of two batoids—a rajid skate (Leucoraja erinacea, little skate) and a myliobatid ray (Rhinoptera bonasus, cownose ray)—to determine whether homologous genetic underpinnings pattern the pectoral fins of these species and to reveal patterns that may be shared among divergent batoid lineages.
Using de novo transcriptome assembly and reciprocal-BLAST annotation with multiple vertebrates including Danio rerio, we confirmed expected gene expression patterns associated with the specific tissue domains dissected in R. bonasus (Figure S2). We then evaluated differential expression of genes in each R. bonasus pectoral fin domain relative to the others (Figure 3; Table S4). We found expression of Hoxd10-12 and Grem1 to be confined to the posterior pectoral fin, while Hoxd9 and Hoxa9 were most highly expressed in the mid-posterior pectoral fin, consistent with canonical patterning of paired appendages in vertebrates (Khokha et al., 2003; Zákány et al., 2004; Freitas et al., 2007; Ahn and Ho, 2008; Figure 3). We found genes associated with posterior patterning of pectoral fins in L. erinacea enriched in the posterior of R. bonasus pectoral fins as well (Figure 4, green dots), confirming that our samples were undergoing similar patterning processes and that the stages were comparable with respect to fin development (though note that the posteriorly expressed Ler-Grem1 transcript did not assemble well and was therefore not detected). Further, we confirmed that expression of Tbx4 and Tbx5, genes which define developing pelvic and pectoral fins in vertebrates, respectively (Gibson-Brown et al., 1998; Ahn et al., 2002), exhibited expected expression profiles in R. bonasus (for example, average counts per million in Tbx4 was >20x higher in the pelvic fin (280) when compared to pectoral fin domains (10); whereas the opposite was true with Tbx5). Taken together, these observations indicate that the cownose ray RNA-Seq data reliably show domain-specificity and indicate that similar processes drive development of the posterior pectoral fins of skates and myliobatids.
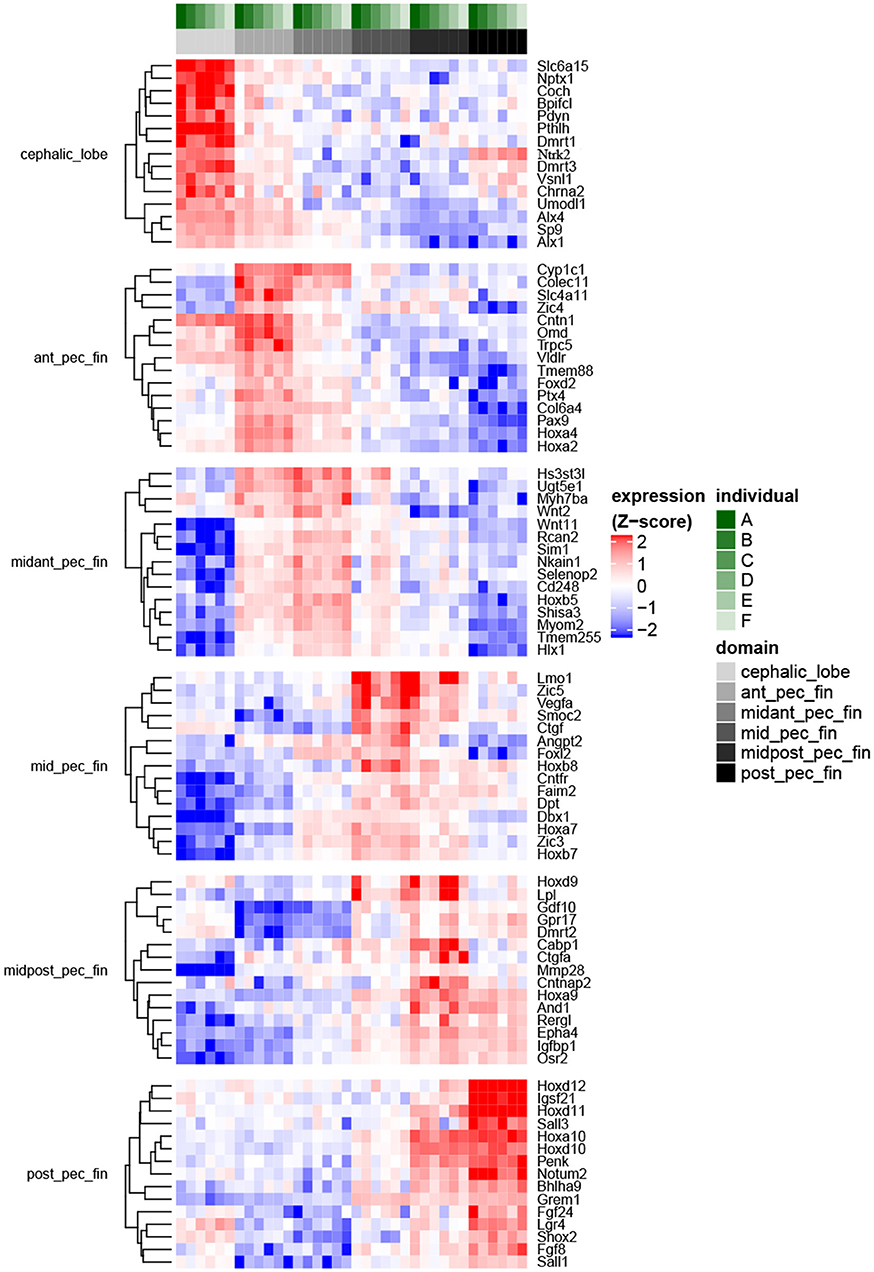
Figure 3. Fin domain-specific expression patterns highlight both ancestral and derived developmental programs in R. bonasus. Differential expression was assessed via the quasi-likelihood f-test (refer to Methods). The heatmap represents genes with transcripts most strongly up-regulated in each of six sampled fin domains when contrasted with the other domains (FDR < 0.001, top 15 genes by log2FC are shown). Transcripts that were up-regulated in more than one domain were assigned to the single domain with the highest expression. Also, transcripts assigned to different isoforms of D. rerio genes (i.e., “a” vs. “b”) were summarized such that only those with the largest expression differences between domains are shown.
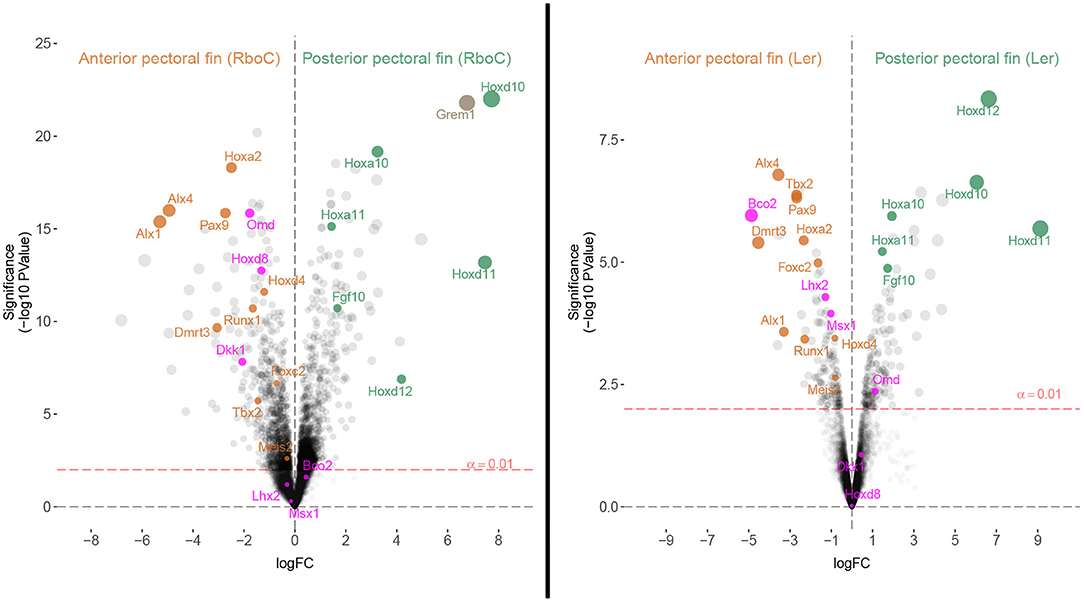
Figure 4. Homologous patterning mechanisms of the anterior and posterior fins of cownose ray and little skate embryos are evident in transcriptomic comparisons. Volcano plots depicting differentially expressed genes in stage 3 R. bonasus embryos and stage 31 L. erinacea embryos. In R. bonasus, both the cephalic lobe and anterior domains (Anterior pectoral fin (RboC)) were contrasted with the posterior and mid-posterior domains (Posterior pectoral fin (RboC); see Figure 1), in order to make comparisons with the anterior and posterior L. erinacea fin domains examined by Nakamura et al. (2015). The left side of each graph represents transcripts biased in the anterior pectoral fin while the right side represents transcripts biased in the posterior pectoral fin. Labeled transcripts include those that are differentially expressed in the anterior pectoral fins of both species (orange), those that are differentially expressed in the posterior pectoral fins of both species (green), and those that have distinct expression patterns in each species (purple). Six replicates for each fin domain were used in R. bonasus comparisons and two anterior vs. three posterior fin replicates were used in L. erinacea comparisons. Transcripts without high quality annotations were removed for clarity, such that only one transcript is reported for each gene.
To evaluate homology between the anterior pectoral fin regions of myliobatid stingrays and rajid skates based on shared expression of anterior patterning genes, we identified genes known to be associated with patterning anterior regions of paired appendages and examined their expression in the anterior pectoral fin region of L. erinacea (anterior third, hereafter APFLer) and the comparable region in R. bonasus (hereafter APFRboC; see Figure 1), which includes the cephalic lobes. By comparing ratios of gene expression in these domains relative to the posterior domains (within each species separately), we identified genes that are similarly enriched in the anterior and posterior pectoral fins of R. bonasus and L. erinacea (Figure 4). These include genes that are known to be associated with anterior patterning in fins and limbs such as Pax9, Alx4, Alx1, and Tbx2 (Onimaru et al., 2015) as well as Foxc2, Runx1, Meis2 and Dmrt3 (Figure 4, orange dots). We also found upregulation of Hoxa2 and Hoxd4 in the anterior pectoral fin regions of both species (Figure 4), which are likely associated with a batoid-specific AER in this region (Nakamura et al., 2015). Taken together, these data support homology of the anterior pectoral fin regions—which includes cephalic lobes in the Myliobatidae—in two disparate batoid species with distinct morphologies.
By sequencing six pectoral fin domains in the cownose ray instead of three, we added a layer of spatial resolution to our analyses which also revealed fine-scale expression patterns that were not apparent when considering broad domains for comparison with the little skate. This experimental design revealed high levels of expression of Hoxd8, Ntrk2, and Enpp2 at the anterior of developing cownose ray fins corresponding to the cephalic lobe (Figure 5; Table S5). In addition, the increased degree of spatial resolution allowed us to find enrichment of Wnt3 and Hoxa13 in the anterior region of R. bonasus pectoral fins (Figure 5), genes which are also enriched in the anterior of L. erinacea pectoral fins as indicated by in-situ hybridization (Nakamura et al., 2015; Barry and Crow, 2017). Wnt3 is associated with a novel apical ectodermal ridge (AER) driving outgrowth at the anterior of the L. erinacea pectoral fin (Nakamura et al., 2015). Hoxa13, a gene involved in patterning novel paired fin domains in batoids (O'Shaughnessy et al., 2015; Barry and Crow, 2017), has overlapping expression with Wnt3, suggesting a role for specification of the anterior AER in batoids (Barry and Crow, 2017). Enrichment of Hoxa13 and Wnt3 in the cephalic lobes of R. bonasus suggests that the anterior AER found in skates is correlated with development of the anterior pectoral fin region (including cephalic lobes) in derived batoids as well. We note that the high expression of these genes in the anterior pectoral fin region of R. bonasus was initially masked (see, for example, Hoxa13 in Figure 5), because they are upregulated in the cephalic lobe (red in Figure 5) but downregulated in the anterior (blue in Figure 5), resulting in signal cancellation when the expression scores for these domains were merged. Importantly, we found no evidence for independent posterior patterning by 5′ HoxD or Shh genes in cephalic lobes, supporting the notion that cephalic lobes do not represent a third set of independent paired appendages, but are instead modified anterior pectoral fins.
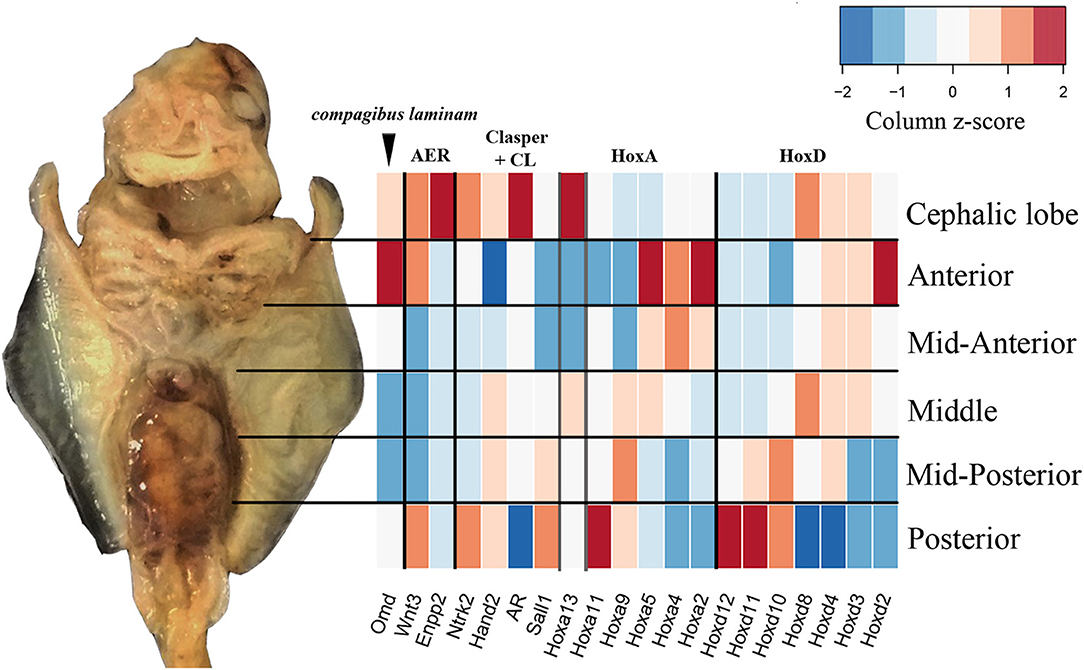
Figure 5. Fine-scale expression patterns are evident during R. bonasus fin development. A representative stage three R. bonasus embryo is shown in ventral view with anterior up. The heatmap represents the average read counts for 6 biological replicates of each fin domain (log2-transformed counts per million, CPM), visualized using a column-based z-score standardization. Gene annotations were originally based on annotation to D. rerio and were confirmed by hand (see Table S3 and Figure S1).
Modified Gene Expression Patterns Associated With Unique Myliobatid Features
Unique morphological features arise in association with changes to developmental processes and gene expression patterns. Therefore, genes that exhibit distinct expression profiles in R. bonasus and L. erinacea during development of the pectoral fins may be associated with morphological differences. We have observed that cephalic lobe development is correlated with at least two surprisingly subtle modifications to the process observed in skate pectoral fins: establishment of a “notch” that defines the cephalic lobe-pectoral fin boundary, and a dorsoventral deviation during pectoral fin fusion. Because pectoral fin fusion is common to both skates and cownose rays, it is difficult to identify genes associated with fusion disparities using our comparative approach. However, we were able to identify candidate notch genes by searching our DGE results for genes that have different expression patterns in the APFRboC and the APFLer relative to the posterior pectoral fin (i.e., up or downregulated in the anterior of one species but not the other, see pink genes in Figure 4). We then searched the literature for functional data related to these genes in other taxa and highlighted those known to be associated with processes that likely underlie development of the notch (e.g., cell proliferation, AER signaling) or the compagibus laminam (e.g., calcification, chondrogenic fusion) in Table 3.
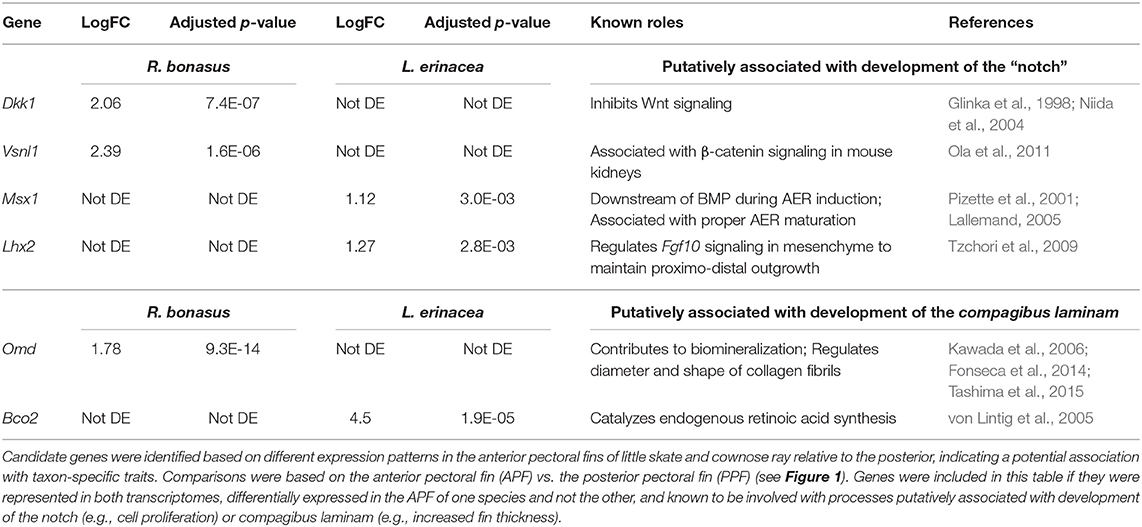
Table 3. Genes that are putatively associated with development of pectoral fin modifications in Myliobatids.
Using these criteria, we identified six genes that may be associated with distinct morphologies at the anterior region of the pectoral fins of R. bonasus and L. erinacea (Table 3). Dkk1 is a known inhibitor of the Wnt signaling pathway (Glinka et al., 1998; Fedi et al., 1999; Niida et al., 2004), while Vsnl1 downregulates β-catenin—an important component of canonical Wnt signaling—in mouse kidneys (Ola et al., 2011). These genes are upregulated in R. bonasus but not L. erinacea and may have a role in notch development by inhibiting AER-signaling in this region. Alternatively, Lhx2 and Msx1 promote AER signaling and appendage outgrowth during development (Pizette et al., 2001; Lallemand, 2005; Tzchori et al., 2009). These genes are enriched at the anterior of L. erinacea and not R. bonasus; downregulation of these genes in R. bonasus may preclude AER-driven outgrowth at the notch. Omd is associated with biomineralization (Kawada et al., 2006; Tashima et al., 2015) and may contribute to development of the thicker fin rays in the compagibus laminam of myliobatids. RNA-Sequencing is a powerful tool for identifying genes that may be associated with novel or modified features during development; however, functional studies would be necessary to validate these associations.
Are Paired Fin Modifications Such as Cephalic Lobes and Claspers Specified by Similar Gene Expression Patterns?
While the CFT pockets and cephalic lobes are fusing at the anterior of the pectoral fins in cownose rays, specification of claspers begins at the posterior pelvic fins where they are distinguished by a notch that is reminiscent of the notch that occurs between the cephalic lobes and pectoral fins (see Figures 2B,D). As the claspers differentiate from the pelvic fins, they assume a paddle shaped morphology (Figures 2D,E) before curling into tubes (Figures 2F,G), transiently resembling the curled resting state of cephalic lobes in Mobula. As both claspers and cephalic lobes are modified fin domains supported by fin rays, the morphological similarities we observed during development led us to ask whether similar molecular processes could underlie development of these features.
In addition to the pectoral fin domains described above, we sequenced the transcriptome of pelvic fins of the six stage three cownose rays. At this stage, clasper morphogenesis has not yet begun, but we were able to diagnose sex based on expression of genes known to be exclusively expressed during clasper development in males (i.e. not expressed in pelvic fin development of females) such as Hoxd12, Hoxd13, Hand2 (O'Shaughnessy et al., 2015) and Hoxa13 (Barry and Crow, 2017). We confirmed statistically significant enrichment of Hoxd13, Hoxa13, and Hand2 in the pelvic fins of three of our samples relative to the others (Table 4), indicating that we had sampled three males and three females. We attributed other differentially expressed genes between male and female pelvic fin domains to clasper development, finding enrichment of Sall1, Cdh23, Sall3, Kit, Six2, and Cyp7a1 in male pelvic fins, adding to the repository of genes likely to be associated with clasper development (but note that Sall1 is expressed in posterior pectoral fins as well, see Figures 3, 5).
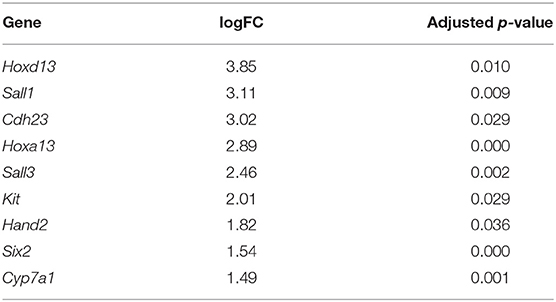
Table 4. Genes differentially expressed in the pelvic fins of R. bonasus males (n = 3) relative to females (n = 3) prior to clasper morphogenesis (adj. p-values < 0.05).
Interestingly, we found enrichment of five genes in both claspers and cephalic lobes, suggesting a redeployment of outgrowth and patterning pathways during development of these paired fin modifications. In addition to their enrichment in male pelvic fins relative to female, Hand2 and Sall1 are enriched in the cephalic lobes relative to the adjacent pectoral fin domains (Table S5). Ntrk2 is similarly upregulated in claspers (logFC = 1.54, p-value = 0.0002, adj. p-value = 0.13) and highly expressed in cephalic lobes (Table S4). Finally, Hoxa13 is expressed specifically in developing claspers of male pelvic fins, and is also expressed in anterior pectoral fins of the little skate (Barry and Crow, 2017) and cownose ray (Figure 5). Each of these patterns was obscured when examining broad domains in the cownose ray due to low expression in the domain adjacent to cephalic lobe. However, our experimental design allowed us to detect enrichment of these genes specifically in the anterior pectoral fin region corresponding to cephalic lobes (Figure 5). Although we did not detect enrichment of putative notch genes (those outlined in Table 3) in the pelvic fins of stage three males, this may simply reflect temporal differences in notch development, as the cephalic lobe notch is clearly defined at stage three, while the pelvic fin notch does not appear until stage four (Figure 2D). Intriguingly, androgen receptor (AR), another gene associated with clasper specification (O'Shaughnessy et al., 2015), was also upregulated in the cephalic lobe relative to every other pectoral fin domain (p < 0.01) (see Figure 5 and Table S5). AR was not represented in the L. erinacea transcriptome, so the possibility remains that AR expression at the anterior pectoral fin may be common among batoids. Future studies examining expression of genes associated with clasper development in the pectoral fins and cephalic lobes/APF of batoids may reveal that these paired fin modifications rely on similar developmental-genetic pathways for specification of novel domains.
Discussion
Manta rays and their relatives (Myliobatidae) have functionally partitioned feeding and swimming behaviors into distinct pectoral fin regions which have been modified to optimize these specific tasks. Here, we have examined the morphological and molecular mechanisms underlying development of these modified fin domains in a representative myliobatid, the cownose ray (Rhinoptera bonasus), with a focus on the evolution and development of cephalic lobes - a derived functional trait unique to myliobatids. We have identified developmental processes that are conserved among batoids as well as novel processes and morphologies that are unique to myliobatids with cephalic lobes. We provide evidence supporting homology of myliobatid cephalic lobes with the anterior pectoral fins of rajid skates and suggest candidate genes that may be associated with development of the notch and the compagibus laminam, two features that are unique to the anterior pectoral fins of myliobatids.
Myliobatid Cephalic Lobes Are Homologous to the Anterior Pectoral Fins of Skates
Batoids are distinguished from other cartilaginous fishes by their disc-shaped pectoral fins, which are dorsoventrally flattened and extend anteriorly to fuse to the rostrum (Last et al., 2016). Species of the family Myliobatidae exhibit pectoral fin modifications during development that result in cephalic lobes. Until this study, the process by which cephalic lobes develop, including their physical attachment to developing pectoral fins, had not been documented. Interestingly, the presence of cephalic lobes is not correlated with an increase in the number of fin rays articulating to the propterygium in myliobatids (Hall et al., 2018), as would be expected if cephalic lobes were novel additions to ancestral anterior pectoral fins. Rather, the morphological observations and developmental gene expression patterns presented here suggest that cephalic lobes of myliobatids are homologous to the anterior pectoral fin of the little skate–a representative batoid lacking cephalic lobes. These data indicate that the mechanisms underlying anterior expansion of the pectoral fins were present in the common ancestor of skates and rays. The primary difference between developing cownose ray cephalic lobes and skate pectoral fins is the presence of a notch that defines cephalic lobes by separating the anterior pectoral fin into distinct domains. This subtle developmental modification is associated with specification of a distinct region - the cephalic lobe - that is optimized for feeding. The evolution of cephalic lobes is likely associated with modifications to the rest of the pectoral fins that permitted the evolution of oscillatory swimming (a.k.a. underwater flight) and adaptation to a pelagic or bentho-pelagic lifestyle in the Myliobatidae.
Molecular Development of Derived Myliobatid Features
While cephalic lobes are homologous with anterior pectoral fins in skates, there are differences in developmental gene expression patterns associated with unique morphological modifications in myliobatid pectoral fins. Comparative transcriptomics using RNA-sequencing is limited in its capacity for identifying genes that are correlated with distinct phenotypes, as it does not include functional information. Nevertheless, by searching the data for genes that are up or down-regulated at the anterior of one species and not the other, we identified candidate genes that may be associated with myliobatid-specific features such as the compagibus laminam and development of the notch (Table 3; Table S6).
The expression pattern of osteomodulin (omd) is particularly intriguing. In R. bonasus, expression of omd is concentrated at the anterior of the pectoral fin, posterior to the cephalic lobe in the region of the compagibus laminam (Figures 3, 5). The pattern differs from L. erinacea, where omd is slightly upregulated at the posterior (Figure 4). Due to its known role in biomineralization and enriched expression in a pectoral fin domain of R. bonasus that exhibits increased calcification, we hypothesize that omd may be associated with development of the compagibus laminam, a plate of nearly fused and highly calcified fin rays at the anterior of myliobatid pectoral fins (Schaefer and Summers, 2005; Hall et al., 2018) which likely contributes to lift generation during oscillatory swimming.
The notch, a developmental feature that maintains a clear delineation between the cephalic lobe and the rest of the pectoral fin, likely develops as a region that is marked for reduced tissue outgrowth, or interruption of the AER, relative to the ancestral state. Once we identified genes with different expression patterns in the anterior pectoral fins of R. bonasus and L. erinacea, we queried this list for genes that have been shown to be associated with developmental pathways and processes that may be pertinent to notch formation (e.g., Wnt signaling, cell proliferation, apoptosis). Of the identified candidate genes, Dkk1, Msx2, and Lhx2 are compelling candidates for notch induction. Dkk1 is a known Wnt inhibitor. Wnt signaling is an important component of AER induction and maintenance (ten Berge et al., 2008). Upregulation of Wnt inhibiting genes at the notch could provide a mechanism by which AER-facilitated outgrowth could be stunted in the notch while continuing in the adjacent regions. Alternatively, downregulation of genes such as Msx1 and Lhx2 that promote AER maintenance could have a similar effect, resulting in reduced cell proliferation (Table 3). Though we did not identify any candidate notch genes with roles in apoptosis, apoptosis could also be a mechanism by which a notch could form in developing fins. If apoptotic pathways were activated in the notch, AER signaling could occur as a continuous strip throughout the anterior pectoral fin (including cephalic lobe), while an increase in cell death at the notch could prevent tissue outgrowth from keeping pace with adjacent areas. Further research into notch development is needed to confirm the underlying mechanisms, perhaps via functional studies that attempt to induce a notch in a more tractable batoid species such as the little skate (Leucoraja erinacea) via ectopic expression or deletion of the genes highlighted in Table 3.
Development of the anterior region of myliobatid pectoral fins appears to involve a unique combination of Hox gene expression and redeployment of posterior modules. Though most HoxD genes exhibit canonical expression patterns in the pectoral fin of R. bonasus (Figure 5), expression of Hoxd8 is conspicuously enriched in the APF/cephalic lobe of R. bonasus but not L. erinacea (Figures 4, 5), suggesting that there may be a myliobatid-specific Hox code at the anterior of the pectoral fin. In addition, Enpp2 (aka autotaxin), is a known target of Hoxa13 (Williams et al., 2005) and may contribute to AER induction in the anterior pectoral fins of batoids (this gene was not represented in the L. erinacea transcriptome), or specification of cephalic lobes in myliobatids. Mirroring the pattern of Enpp2, Ntrk2 expression is enriched in both the cephalic lobes and the posterior pectoral fins of R. bonasus, as are Wnt3 and Hand2 (Figure 5). By revealing genes with expression patterns that are mirrored at the anterior and posterior of R. bonasus pectoral fins, our domain-specific data suggest that these regions share a handful of common patterning mechanisms, which may contribute to development of boundaries in paired fins.
Developmental Variations in Batoids
Modifications to outgrowth and fusion processes may contribute to the morphological variation observed at the anterior pectoral fins among myliobatid taxa. For example, the single fused cephalic lobe of mature bat rays is found in the same plane as the pectoral fin, indicating that the observed deviation during pectoral fin fusion in the region of the notch in cownose rays is lacking in bat rays (Figures 6A,B). This appears to be due to a dorsoventral shift in the pattern of fusion at the notch region in cownose rays and other myliobatid species. While species in the myliobatid genera Rhinoptera, Mobula, and Aetobatus lack skeletal elements in the shoulder (corresponding to the notch), some bat rays of the genus Myliobatis display a radiation of stunted fin rays in this region between the cephalic lobe and pectoral fin (Figures 6C,D). We hypothesize that, relative to other myliobatids, these bat rays exhibit weak or temporally constricted expression of the genes responsible for suppressing tissue outgrowth in the notch. Combined, these observations suggest that the molecular processes driving notch formation and the pattern of fusion may vary among myliobatid taxa, with representation of intermediate phenotypes.
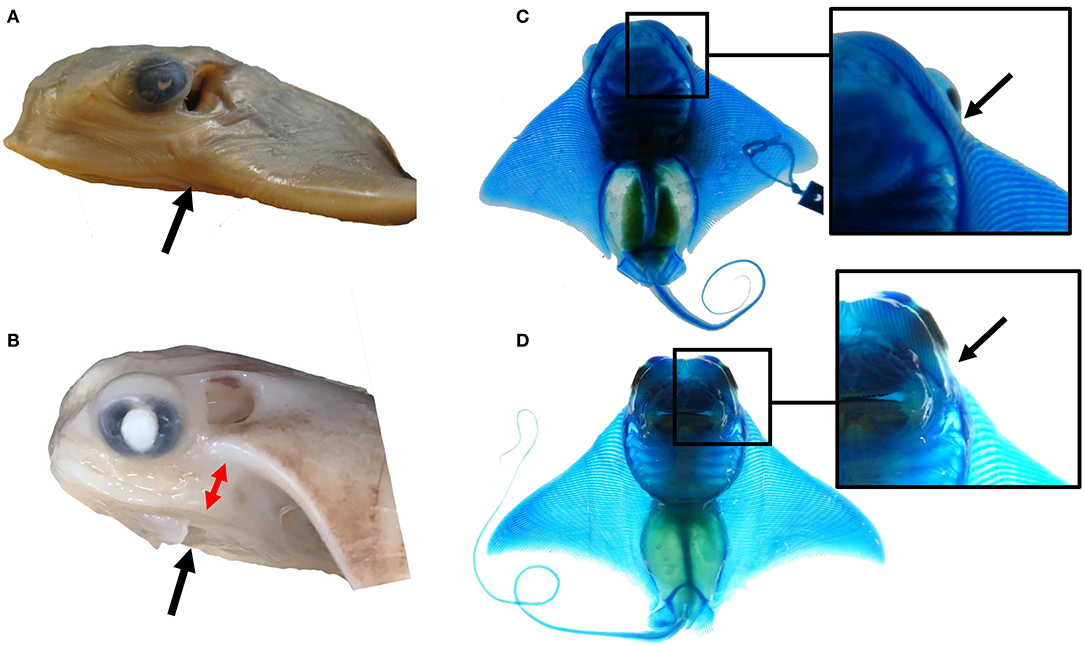
Figure 6. The notch and pattern of pectoral fusion may underlie morphological differences among myliobatid taxa at the anterior of the pectoral fins. (A) The left side of the head of Myliobatis californica, (B) the left side of the head of Rhinoptera bonasus. Black arrows point to the notch domain. Notice the dorso-ventral displacement between the cephalic lobe and the rest of the pectoral fin at this region in R. bonasus (denoted by the red arrows) but not in M. californica. (C) The ventral side of a cleared and stained M. californica specimen, (D) the ventral side of a cleared and stained R. bonasus specimen. Notice the presence of fin rays in the shoulder region of M. californica and their absence in R. bonasus. This region corresponds to the notch during development; variation in skeletal structure here may imply that mechanisms that inhibit tissue outgrowth at the notch are less active in M. californica than R. bonasus.
Aberrant phenotypes reflecting incomplete pectoral fin fusion have been observed in various batoid clades, including the Myliobatidae (Ramírez-Amaro et al., 2013), Rajidae (Prado et al., 2008), Torpedinidae (Mnasri et al., 2010), Dasyatidae (Prado et al., 2008; Ramírez-Hernandez et al., 2011), Urotrygonidae (Mejía-Falla et al., 2011), Potamotrygonidae (Oldfield, 2005a,b), Rhinobatidae (Bornatowski and Abilhoa, 2009), and Gymnuridae (Narváez and Osaer, 2016). This prevalent deformity was first described over a century ago (Gill, 1896) and has since been termed the “batman” morphology (Oldfield 2005a; Oldfield 2005b). After observing that a two-step process consisting of distinct phases of outgrowth and fusion underlies development of the anterior pectoral fins in cownose rays, we suggest that the batman morphology may occur when the molecular mechanisms driving outgrowth proceed normally, while the mechanisms underlying fusion are interrupted. It has been suggested that the batman morphology may be associated with exposure to pollutants (Heupel et al., 1999), which could disrupt developmental-genetic processes (Tomanek, 2011; Varotto et al., 2013).
Are Paired Fin Modifications Such as Cephalic Lobes and Claspers Specified by Similar Gene Expression Patterns?
Claspers are male reproductive organs in cartilaginous fishes, which are modifications to the posterior pelvic fins in extant taxa. Morphologically, clasper formation is preceded by development of a notch between the budding claspers and pelvic fins, reminiscent of the notch that develops between the cephalic lobes and the rest of the pectoral fins. This led us to wonder if specification of cephalic lobes in anterior pectoral fins represents a mirror image of clasper specification in posterior pelvic fins, and if we could find evidence for co-option of this ancient developmental pathway in other modified fin domains.
Transcriptomic data revealed that genes known to be involved in clasper specification are indeed enriched in developing cephalic lobes. Barry and Crow (2017) found that Hoxa13 is expressed exclusively in the claspers of the little skate, and not expressed in female pelvic fins. Variation in the pattern of Hoxa13 expression is often associated with the evolution of novel features (Crow et al., 2009; Archambeault et al., 2014; Barry and Crow, 2017), including the autopod (Sordino et al., 1995; Yokouchi et al., 1995; Fromental-Ramain et al., 1996; Metscher et al., 2005), making it a likely candidate for specification of derived batoid features as well. During clasper development, Hand2 is regulated by hormonal input via upregulation of AR (O'Shaughnessy et al., 2015); it was suggested that the evolution of claspers was facilitated by hormonal input to fin developmental networks following the evolution of androgen response elements at the Hand2 locus (O'Shaughnessy et al., 2015). Interestingly, both Hand2 and AR are similarly enriched in developing cephalic lobes. In conjunction with high levels of Ntrk2 and Sall1 expression in both claspers and cephalic lobes (relative to the adjacent fin domain; see Figure 5, Table S5), our data support an intriguing hypothesis: namely, that cephalic lobes, which are anterior pectoral fin modifications, may have evolved by co-opting developmental pathways associated with the evolution of claspers - posterior pelvic fin modifications in male cartilaginous fishes.
Conclusions
We have used interspecies comparative transcriptomics to shed light on the evolution and development of paired fins in a batoid family with derived features (Myliobatidae). We found that myliobatid pectoral fins develop via a two-step process in which they first extend anterodistally and subsequently fuse lateral to the gill arches and rostrum, while cephalic lobes develop as modified anterior pectoral fins that fuse anterior to the mouth. Morphologically, the feature that distinguishes the myliobatid pectoral fin from that of other batoids during early fin development is the notch, while ventral deviation during fusion of the pectoral fins may be unique to derived myliobatid taxa with ventrally-positioned cephalic lobes. We find no evidence supporting cephalic lobes as independent paired appendages; rather, cephalic lobes are modified anterior pectoral fin domains that share homology with the anterior pectoral fins of skates while also exhibiting distinct expression patterns of a handful of candidate genes, including some that are known to be associated with clasper development. This research represents a foray into the frontiers of evolutionary developmental biology: by applying advanced molecular techniques to elucidate the evolution and development of a non-model organism with derived functional traits, we have learned that the distinctive myliobatid body plan evolved via subtle changes to ancestral developmental pathways.
Data Availability
Transcriptome sequencing data have been deposited at the National Center for Biotechnology Information Sequence Read Archives (NCBI SRA), www.ncbi.nlm.nih.gov/sra (BioProject accession code PRJNA497831). Assembled transcriptomes and annotation files have been added to Figshare (doi: 10.6084/m9.figshare.7265579).
Ethics Statement
This study was carried out in accordance with the recommendations of the San Francisco State University Institutional Animal Care and Use Committee (IACUC) under protocol A15-09R2. The protocol was approved by the IACUC.
Author Contributions
JS and KC designed research and wrote manuscript. JS and RF collected samples and field data. JS performed labwork. JK assembled transcriptome and performed DGE analyses. All authors edited and approved final manuscript.
Funding
Myers Ocean Trust, American Elasmobranch Society, Sigma XI Grants-In-Aid-of-Research, CSUPERB travel grant, COAST graduate student research award, SFSU IRA and GSCB research awards to JS; SFSU MiSeq-Mini Grant support from National Science Foundation (NSF) award 1427772; and NSF Award Number: IOS 1656487 to KC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are tremendously grateful to George Trice Sr., George Trice Jr., Tommy Lewis, Dimitri Hionis, and Christine Bedore for assistance with sample collection; Gregg Poulakis, Rachel Scharer, and the Florida Fish and Wildlife Conservation Commission for insight into the logistics of fieldwork and conversations that helped move this project forward; Dave Catania and the California Academy of Sciences Ichthyology collection for loaning specimens of Myliobatis californica; Kayla Hall for clearing, staining, and photographing the cownose rays and bat rays used in Figure 6; Tetsuya Nakamura for sharing little skate RNA-Seq data; UC Berkeley Functional Genomics Laboratory and Vincent J. Coates Genomics Sequencing Laboratory for quality assessment of samples and sequencing; Michelle Conrad and Frank Cipriano for help with additional quality assessment and sequencing at San Francisco State University; and the UC Davis Bioinformatics Core for initial guidance with data processing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00181/full#supplementary-material
References
Agerman, K. (2003). BDNF gene replacement reveals multiple mechanisms for establishing neurotrophin specificity during sensory nervous system development. Development 130, 1479–1491. doi: 10.1242/dev.00378
Ahn, D., and Ho, R. K. (2008). Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages. Dev. Biol. 322, 220–233. doi: 10.1016/j.ydbio.2008.06.032
Ahn, D. G., Kourakis, M. J., Rohde, L. A., Silver, L. M., and Ho, R. K. (2002). T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature 417, 754–758. doi: 10.1038/nature00814
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andrews, S. (2010). FastQC. Babraham Bioinformatics. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Archambeault, S., Taylor, J. A., and Crow, K. D. (2014). HoxA and HoxD expression in a variety of vertebrate body plan features reveals an ancient origin for the distal Hox program. EvoDevo 5:44. doi: 10.1186/2041-9139-5-44
Ari, C., and Correia, J. P. (2008). Role of sensory cues on food searching behavior of a captive Manta birostris (Chondrichtyes, Mobulidae). Zoo Biol. 27, 294–304. doi: 10.1002/zoo.20189
Aschliman, N. C. (2014). Interrelationships of the durophagous stingrays (Batoidea: Myliobatidae). Environ. Biol. Fishes 97, 967–979. doi: 10.1007/s10641-014-0261-8
Aschliman, N. C., Nishida, M., Miya, M., Inoue, J. G., Rosana, K. M., and Naylor, G. J. (2012). Body plan convergence in the evolution of skates and rays (Chondrichthyes: Batoidea). Mol. Phylogenet. Evol. 63, 28–42. doi: 10.1016/j.ympev.2011.12.012
Babel, J. S. (1966). Fish Bulletin 137. Reproduction, Life History, and Ecology of the Round Stingray, Urolophus Halleri Cooper. San Diego, CA: Library – Scripps Collection. Retrieved from: https://escholarship.org/uc/item/79v683zd
Ballard, W. W., Mellinger, J., and Lechenault, H. (1993). A series of normal stages for development of Scyliorhinus canicula, the lesser spotted dogfish (Chondrichthyes: Scyliorhinidae). J. Exp. Zool. 267, 318–336. doi: 10.1002/jez.1402670309
Barry, S. N., and Crow, K. D. (2017). The role of HoxA11 and HoxA13 in the evolution of novel fin morphologies in a representative batoid (Leucoraja erinacea). EvoDevo 8:24. doi: 10.1186/s13227-017-0088-4
Bedore, C. N., Harris, L. L., and Kajiura, S. M. (2014). Behavioral responses of batoid elasmobranchs to prey-simulating electric fields are correlated to peripheral sensory morphology and ecology. Zoology 117, 95–103. doi: 10.1016/j.zool.2013.09.002
Bornatowski, H., and Abilhoa, V. (2009). Record of an anomalous embryo of Rhinobatos percellens (Elasmobranchii: Rhinobatidae) in the southern coast of Brazil. Mar. Biodivers. Rec. 2:e36. doi: 10.1017/S1755267209000414
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chang, D. T., López, A., Kessler, D. P., von Chiang, C., Simandl, B. K., Zhao, R., et al. (1994). Products, genetic linkage and limb patterning activity of a murine hedgehog gene. Development 120, 3339–3353.
Couturier, L. I., Marshall, A. D., Jaine, F. R. A., Kashiwagi, T., Pierce, S. J., Townsend, K. A., et al. (2012). Biology, ecology and conservation of the Mobulidae. J. Fish Biol. 80, 1075–1119. doi: 10.1111/j.1095-8649.2012.03264.x
Crow, K. D., Amemiya, C. T., Roth, J., and Wagner, G. P. (2009). Hypermutability of Hoxa13a and functional divergence from its paralog are associated with the origin of a novel developmental feature in zebrafish and related taxa (Cypriniformes). Evolution 63, 1574–1592. doi: 10.1111/j.1558-5646.2009.00657.x
Dahn, R. D., Davis, M. C., Pappano, W. N., and Shubin, N. H. (2007). Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature 445, 311–314. doi: 10.1038/nature05436
Dunn, C. W., Luo, X., and Wu, Z. (2013). Phylogenetic analysis of gene expression. Integr. Comp. Biol. 53, 847–856. doi: 10.1093/icb/ict068
Fedi, P., Bafico, A., Soria, A. N., Burgess, W. H., Miki, T., Bottaro, D. P., et al. (1999). Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J. Biol. Chem. 274, 19465–19472. doi: 10.1074/jbc.274.27.19465
Fisher, R. A., Call, G. C., and Grubbs, R. D. (2011). Cownose ray (Rhinoptera bonasus) predation relative to bivalve ontogeny. J. Shellfish Res. 30, 187–196. doi: 10.2983/035.030.0126
Fonseca, H., Moreira-Gonçalves, D., Coriolano, H. J., and Duarte, J. A. (2014). Bone quality: the determinants of bone strength and fragility. Sports Med. 44, 37–53. doi: 10.1007/s40279-013-0100-7
Fontanella, J. E., Fish, F. E., Barchi, E. I., Campbell-Malone, R., Nichols, R. H., DiNenno, N. K., et al. (2013). Two- and three-dimensional geometries of batoids in relation to locomotor mode. J. Exp. Mar. Biol. Ecol. 446, 273–281. doi: 10.1016/j.jembe.2013.05.016
Franklin, O., Palmer, C., and Dyke, G. (2014). Pectoral fin morphology of batoid fishes (Chondrichthyes: Batoidea): explaining phylogenetic variation with geometric morphometrics. J. Morphol. 275, 1173–1186. doi: 10.1002/jmor.20294
Freitas, R., Gómez-Marín, C., Wilson, J. M., Casares, F., and Gómez-Skarmeta, J. L. (2012). Hoxd13 contribution to the evolution of vertebrate appendages. Dev. Cell 23, 1219–1229. doi: 10.1016/j.devcel.2012.10.015
Freitas, R., Zhang, G., and Cohn, M. J. (2007). Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLoS ONE 2:e754. doi: 10.1371/journal.pone.0000754
Fromental-Ramain, C., Warot, X., Messadecq, N., LeMeur, M., Dollé, P., and Chambon, P. (1996). Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997–3011.
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y., Gentry, J., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. doi: 10.1186/gb-2004-5-10-r80
Gibson-Brown, J. J., Agulnik, S. I., Silver, L. M., Niswander, L., and Papaioannou, V. E. (1998). Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development 125, 2499–2509.
Gill, T. (1896). Notes on the genus Cephaleutherus of Rafinesque, and other rays with aberrant pectoral fins (Propterygia and Hieroptera). Proc. U. S. Natl. Mus. 18, 195–198. doi: 10.5479/si.00963801.18-1054.195
Glinka, A., Wu, W., Delius, H., Monaghan, A. P., Blumenstock, C., and Niehrs, C. (1998). Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391, 357–362. doi: 10.1038/34848
Hall, K. C., Hundt, P. J., Swenson, J. D., Summers, A. P., and Crow, K. D. (2018). The evolution of underwater flight: the redistribution of pectoral fin rays, in manta rays and their relatives (Myliobatidae). J. Morphol. 279, 1155–1170. doi: 10.1002/jmor.20837
Heupel, M. R., Simpfendorfer, C. A., and Bennett, M. B. (1999). Skeletal deformities in elasmobranchs from Australian waters. J. Fish Biol. 54, 1111–1115. doi: 10.1111/j.1095-8649.1999.tb00861.x
Ihaka, R., and Gentleman, R. (1996). R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314.
Inoue, J. G., Miya, M., Lam, K., Tay, B. H., Danks, J. A., Bell, J., et al. (2010). Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol. Biol. Evol. 27, 2576–2586. doi: 10.1093/molbev/msq147
Jordan, L. K. (2008). Comparative morphology of stingray lateral line canal and electrosensory systems. J. Morphol. 269, 1325–1339. doi: 10.1002/jmor.10660
Kawada, M., Hamajima, S., and Abiko, Y. (2006). Osteomodulin gene expression in human bone marrow-derived mesenchymal stem cells into osteoblastic differentiation. Int. J. Oral-Med. Sci. 4, 170–176. doi: 10.5466/ijoms.4.170
Khokha, M. K., Hsu, D., Brunet, L. J., Dionne, M. S., and Harland, R. M. (2003). Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 34, 303–307. doi: 10.1038/ng1178
Kmita, M., Tarchini, B., Zàkàny, J., Logan, M., Tabin, C. J., and Duboule, D. (2005). Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435, 1113–1116. doi: 10.1038/nature03648
Krauss, S., Concordet, J. P., and Ingham, P. W. (1993). A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75, 1431–1444. doi: 10.1016/0092-8674(93)90628-4
Krueger, F. (2012). Trim Galore! Babraham Bioinformatics. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
Krumlauf, R. (1994). Hox genes in vertebrate development. Cell 78, 191–201. doi: 10.1016/0092-8674(94)90290-9
Lallemand, Y. (2005). Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development 132, 3003–3014. doi: 10.1242/dev.01877
Last, P., Naylor, G., Séret, B., White, W., Stehmann, M., and Carvalho, M. de. (2016). Rays of the World. Clayton South, VIC: Csiro Publishing.
Liao, Y., Smyth, G. K., and Shi, W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. doi: 10.1093/nar/gkt214
Luer, C. A., Walsh, C. J., Bodine, A. B., and Wyffels, J. T. (2007). Normal embryonic development in the clearnose skate, Raja eglanteria, with experimental observations on artificial insemination. Environ. Biol. Fishes 80, 239–255. doi: 10.1007/s10641-007-9219-4
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Martinez, C. M., Rohlf, F. J., and Frisk, M. G. (2016). Re-evaluation of batoid pectoral morphology reveals novel patterns of diversity among major lineages. J. Morphol. 277, 482–493. doi: 10.1002/jmor.20513
Maxwell, E. E., Fröbisch, N. B., and Heppleston, A. C. (2008). Variability and conservation in late chondrichthyan development: ontogeny of the Winter Skate (Leucoraja ocellata). Anat. Rec. Adv. Integr. Anat. Evol. Biol. 291, 1079–1087. doi: 10.1002/ar.20719
Mejía-Falla, P. A., Navia, A. F., and Muñoz, L. A. (2011). First record of morphological abnormality in embryos of Urotrygon rogersi (Jordan & Starks, 1895) (Myliobatiformes: Urotrygonidae) in the Tropical Eastern Pacific. Lat. Am. J. Aquat. Res. Valpso. 39, 184–188. doi: 10.3856/vol39-issue1-fulltext-19
Metscher, B. D., Takahashi, K., Crow, K., Amemiya, C., Nonaka, D. F., and Wagner, G. P. (2005). Expression of Hoxa-11 and Hoxa-13 in the pectoral fin of a basal ray-finned fish, Polyodon spathula: implications for the origin of tetrapod limbs. Evol. Dev. 7, 186–195. doi: 10.1111/j.1525-142X.2005.05021.x
Miyake, T., McEachran, J. D., Walton, P. J., and Hall, B. K. (1992). Development and morphology of rostral cartilages in batoid fishes (Chondrichthyes: Batoidea), with comments on homology within vertebrates. Biol. J. Linn. Soc. 46, 259–298. doi: 10.1111/j.1095-8312.1992.tb00864.x
Mnasri, N., El Kamel, O., Boumaïza, M., Ben Amor, M. M., Reynaud, C., and Capapé, C. (2010). Morphological abnormalities in two batoid species (Chondrichthyes) from northern Tunisian waters (central Mediterranean). Ann. Ser. Hist. Nat. 20, 181–190.
Morgan, M., Pagès, H., Obenchain, V., and Hayden, N. (2018). Rsamtools: Binary Alignment (BAM), FASTA, Variant Call (BCF), and Tabix File Import. R package version 1.32.0. Available online at: http://bioconductor.org/packages/release/bioc/html/Rsamtools.html
Mulvany, S., and Motta, P. J. (2013). The morphology of the cephalic lobes and anterior pectoral fins in six species of batoids: batoid pectoral fin anatomy. J. Morphol. 274, 1070–1083. doi: 10.1002/jmor.20163
Mulvany, S., and Motta, P. J. (2014). Prey capture kinematics in batoids using different prey types: investigating the role of the cephalic lobes. J. Exp. Zool. Part Ecol. Genet. Physiol. 321, 515–530. doi: 10.1002/jez.1883
Nakamura, T., Klomp, J., Pieretti, J., Schneider, I., Gehrke, A. R., and Shubin, N. H. (2015). Molecular mechanisms underlying the exceptional adaptations of batoid fins. Proc. Natl. Acad. Sci. 112, 15940–15945. doi: 10.1073/pnas.1521818112
Narváez, K., and Osaer, F. (2016). Morphological and functional abnormality in the spiny butterfly ray Gymnura altavela. Mar. Biodivers. Rec. 9:95. doi: 10.1186/s41200-016-0085-7
Nelson, J. S., Grande, T. C., and Wilson, M. V. H. (2016). Fishes of the World. New York, NY: John Wiley & Sons.
Niida, A., Hiroko, T., Kasai, M., Furukawa, Y., Nakamura, Y., Suzuki, Y., et al. (2004). DKK1, a negative regulator of Wnt signaling, is a target of the β-catenin/TCF pathway. Oncogene 23, 8520–8526. doi: 10.1038/sj.onc.1207892
Nishida, K. (1990). Phylogeny of the suborder Myliobatidoidei. Mem. Fac. Fish. Hokkaido Univ. Jpn. 37, 1–108.
Notarbartolo-Di-Sciara, G. (1987). A revisionary study of the genus Mobula Rafinesque, 1810 (Chondrichthyes: Mobulidae) with the description of a new species. Zool. J. Linn. Soc. 91, 1–91. doi: 10.1111/j.1096-3642.1987.tb01723.x
Notarbartolo-di-Sciara, G., and Hillyer, E. V. (1989). Mobulid rays off Eastern Venezuela (Chondrichthyes, Mobulidae). Copeia 1989, 607–614. doi: 10.2307/1445487
Ola, R., Jakobson, M., Kvist, J., Perälä, N., Kuure, S., Braunewell, K. -H., et al. (2011). The Gdnf farget Vsnl1 marks the ureteric tip. J. Am. Soc. Nephrol. 22, 274–284. doi: 10.1681/ASN.2010030316
Oldfield, R. G. (2005a). Biology, husbandry, and reproduction of freshwater stingrays I. Trop. Fish Hobbyist. 53, 114–116.
Oldfield, R. G. (2005b). Biology, husbandry, and reproduction of freshwater stingrays II. Trop. Fish Hobbyist. 54, 110–112.
Onimaru, K., Kuraku, S., Takagi, W., Hyodo, S., Sharpe, J., and Tanaka, M. (2015). A shift in anterior–posterior positional information underlies the fin-to-limb evolution. eLife 4:e07048. doi: 10.7554/eLife.07048
Onimaru, K., Motone, F., Kiyatake, I., Nishida, K., and Kuraku, S. (2018). A staging table for the embryonic development of the brownbanded bamboo shark (Chiloscyllium punctatum). Dev. Dyn. 247, 712–723. doi: 10.1002/dvdy.24623
O'Shaughnessy, K. L., Dahn, R. D., and Cohn, M. J. (2015). Molecular development of chondrichthyan claspers and the evolution of copulatory organs. Nat. Commun. 6:6698. doi: 10.1038/ncomms7698
Pita-Thomas, W., Fernández-Martos, C., Yunta, M., Maza, R. M., Navarro-Ruiz, R., Lopez-Rodríguez, M. J., et al. (2010). Gene expression of axon growth promoting factors in the deer antler. PLoS ONE 5:e15706. doi: 10.1371/journal.pone.0015706
Pizette, S., Abate-Shen, C., and Niswander, L. (2001). BMP controls proximodistal outgrowth, via induction of the apical ectodermal ridge, and dorsoventral patterning in the vertebrate limb. Development 128, 4463–4474.
Poortvliet, M., Olsen, J. L., Croll, D. A., Bernardi, G., Newton, K., Kollias, S., et al. (2015). A dated molecular phylogeny of manta and devil rays (Mobulidae) based on mitogenome and nuclear sequences. Mol. Phylogenet. Evol. 83, 72–85. doi: 10.1016/j.ympev.2014.10.012
Prado, C. C. R., Oddone, M. C., Gonzalez, M. M. B., de Amorim, A. F., and Capapé, C. (2008). Morphological abnormalities in skates and rays (Chondrichthyes) from off southeastern Brazil. Arq. Ciênc. Mar. 41, 21–28. doi: 10.32360/acmar.v41i2.6058
Ramírez-Amaro, S. R., González-Barba, G., Galván-Magaña, F., and Cartamil, D. (2013). First record of abnormal cephalic horns in the California bat ray Myliobatis californica. Mar. Biodivers. Rec. 6:e24. doi: 10.1017/S1755267213000146
Ramírez-Hernandez, A., Palacios-Barreto, P., Diego Gaitan-Espitia, J., Reyes, F., and Ramírez, J. (2011). Morphological abnormality in the longnose stingray Dasyatis guttata (Myliobatiformes: Dasyatidae) in the Colombian Caribbean. Anat. Rec. 251, 417–430.
Riddle, R. D., Johnson, R. L., Laufer, E., and Tabin, C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416. doi: 10.1016/0092-8674(93)90626-2
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. doi: 10.1186/gb-2010-11-3-r25
Rosenberger, L. J. (2001). Pectoral fin locomotion in batoid fishes: undulation versus oscillation. J. Exp. Biol. 204, 379–394.
Sasko, D. E., Dean, M. N., Motta, P. J., and Hueter, R. E. (2006). Prey capture behavior and kinematics of the Atlantic cownose ray, Rhinoptera bonasus. Zoology 109, 171–181. doi: 10.1016/j.zool.2005.12.005
Schaefer, J. T., and Summers, A. P. (2005). Batoid wing skeletal structure: novel morphologies, mechanical implications, and phylogenetic patterns. J. Morphol. 264, 298–313. doi: 10.1002/jmor.10331
Simpson, J. T., Wong, K., Jackman, S. D., Schein, J. E., Jones, S. J., and Birol, I. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123. doi: 10.1101/gr.089532.108
Sordino, P., van der Hoeven, F., and Duboule, D. (1995). Hox gene expression in teleost fins and the origin of vertebrate digits. Nat. Lond. 375, 678–681. doi: 10.1038/375678a0
Tashima, T., Nagatoishi, S., Sagara, H., Ohnuma, S., and Tsumoto, K. (2015). Osteomodulin regulates diameter and alters shape of collagen fibrils. Biochem. Biophys. Res. Commun. 463, 292–296. doi: 10.1016/j.bbrc.2015.05.053
ten Berge, D., Brugmann, S. A., Helms, J. A., and Nusse, R. (2008). Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135, 3247–3257. doi: 10.1242/dev.023176
Tomanek, L. (2011). Environmental proteomics: changes in the proteome of marine organisms in response to environmental stress, pollutants, infection, symbiosis, and development. Annu. Rev. Mar. Sci. 3, 373–399. doi: 10.1146/annurev-marine-120709-142729
Tzchori, I., Day, T. F., Carolan, P. J., Zhao, Y., Wassif, C. A., Li, L., Lewandoski, M., Gorivodsky, M., Love, P. E., Porter, F. D., et al. (2009). LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136, 1375–1385. doi: 10.1242/dev.026476
van Dijk, E. L., Jaszczyszyn, Y., and Thermes, C. (2014). Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 322, 12–20. doi: 10.1016/j.yexcr.2014.01.008
Varotto, L., Domeneghetti, S., Rosani, U., Manfrin, C., Cajaraville, M. P., Raccanelli, S., et al. (2013). DNA damage and transcriptional changes in the gills of Mytilus galloprovincialis exposed to nanomolar doses of combined metal salts (Cd, Cu, Hg). PLoS ONE 8:e54602. doi: 10.1371/journal.pone.0054602
Venkatesh, B., Lee, A. P., Ravi, V., Maurya, A. K., Lian, M. M., Swann, J. B., et al. (2014). Elephant shark genome provides unique insights into gnathostome evolution. Nature 505, 174–179. doi: 10.1038/nature12826
Vijay, N., Poelstra, J. W., Künstner, A., and Wolf, J. B. (2013). Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol. Ecol. 22, 620–634. doi: 10.1111/mec.12014
von Lintig, J., Hessel, S., Isken, A., Kiefer, C., Lampert, J. M., Voolstra, O., et al. (2005). Towards a better understanding of carotenoid metabolism in animals. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 1740, 122–131. doi: 10.1016/j.bbadis.2004.11.010
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
White, W. T., Corrigan, S., Yang, L., Henderson, A. C., Bazinet, A. L., Swofford, D. L., et al. (2017). Phylogeny of the manta and devilrays (Chondrichthyes: Mobulidae), with an updated taxonomic arrangement for the family. Zool. J. Linn. Soc. 182, 50–75. doi: 10.1093/zoolinnean/zlx018
Wilga, C. D., Maia, A., Nauwelaerts, S., and Lauder, G. V. (2012). Prey handling using whole-body fluid dynamics in batoids. Zoology 115, 47–57. doi: 10.1016/j.zool.2011.09.002
Williams, T. M., Williams, M. E., Kuick, R., Misek, D., McDonagh, K., Hanash, S., et al. (2005). Candidate downstream regulated genes of Hox group 13 transcription factors with and without monomeric DNA binding capability. Dev. Biol. 279, 462–480. doi: 10.1016/j.ydbio.2004.12.015
Yokouchi, Y., Nakazato, S., Yamamoto, M., Goto, Y., Kameda, T., Iba, H., et al. (1995). Misexpression of Hoxa-13 induces cartilage homeotic transformation and changes cell adhesiveness in chick limb buds. Genes Dev. 9, 2509–2522. doi: 10.1101/gad.9.20.2509
Keywords: manta ray, batoid, myliobatid, cephalic lobes, RNA-seq, gene expression, fin development, comparative transcriptomics
Citation: Swenson JD, Klomp J, Fisher RA and Crow KD (2018) How the Devil Ray Got Its Horns: The Evolution and Development of Cephalic Lobes in Myliobatid Stingrays (Batoidea: Myliobatidae). Front. Ecol. Evol. 6:181. doi: 10.3389/fevo.2018.00181
Received: 08 September 2018; Accepted: 23 October 2018;
Published: 13 November 2018.
Edited by:
Alessandro Minelli, Università degli Studi di Padova, ItalyReviewed by:
Paolo Sordino, Stazione Zoologica Anton Dohrn, ItalyShigehiro Kuraku, RIKEN Center for Biosystems Dynamics Research, Japan
Renata Freitas, i3S, Instituto de Investigação e Inovação em Saúde, Portugal
Copyright © 2018 Swenson, Klomp, Fisher and Crow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John D. Swenson, jswenson@umass.edu
 John D. Swenson
John D. Swenson Jeff Klomp
Jeff Klomp Robert A. Fisher3
Robert A. Fisher3  Karen D. Crow
Karen D. Crow