Introduction: Facile and reliable analytical methods to discriminate single nucleotide polymorphisms (SNPs) are of great importance for the realization of personalized medicine. We have already reported a colorimetric SNP genotyping method using salt-induced, non-crosslinking aggregation of double-stranded DNA-modified gold nanoparticles (dsDNA-GNPs)[1]. To develop a more reliable method, the present study exploited a large difference in colloidal stability of dsDNA-GNPs with and without a dangling end is described (Figure 1).
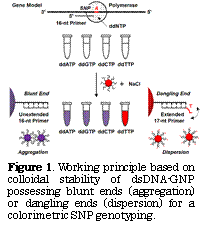
Experimental: According to a reported procedure[1], GNP with a diameter of 15 nm was functionalized with 16-nucleotide (nt) ssDNA. The base sequence of ssDNA was designed to be complementary to an SNP typing primer. A chemically synthesized fragment (56 base pairs) of the cytochrome P450 2C19*2 gene including the SNP site (G→A) was used as a model target. Single base extension of the 16-nt typing primer for the SNP site was carried out with the model target, DNA polymerase, and each ddNTP. To this reaction solution was added ssDNA-GNP (final conc. 5 nM) and NaCl (final conc. 1 M). The as-prepared mixture was incubated for 10 min at room temperature to induce non-crosslinking aggregation. The color change attributed to a surface plasmon resonance shift was detected with the naked eye.
Results and Discussion: When the single base extension reaction using the model target of a homozygous mutant (the SNP site: T) in the presence of ddATP, ddGTP, or ddCTP, the distinct color change from red to purple through non-crosslinking aggregation of dsDNA-GNP was observed (Figure 1, left). This is due to the formation of fully matched dsDNA between ssDNA on the GNP surface and the unextended 16-nt typing primer. On the other hand, it showed no color change (red) with ddTTP (Figure 1, right), because the dsDNA having a dangling end was formed on the surface with the single-base-extended 17-nt typing primer, leading to the nanoparticle’s dispersion by the entropic repulsion. In addition, the corresponding color changes were also obtained when using the model target of the homozygous wild-type gene, as well as that of the heterozygous gene[2].
Conclusion: Colorimetric SNP genotyping of the cytochrome P450 2C19*2 gene by unique colloidal stability of dsDNA-GNP with a dangling end was successfully demonstrated. It will be potentially achievable to develop a clinical sample analysis.
This work was supported by a Grant-in-Aid for Scientific Research (S) (No. 25220204) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by a Grant for Molecular Systems Research provided by RIKEN.
References:
[1] K. Sato, K. Hosokawa, and M. Maeda, Nucleic Acids Res. 2005, 33, e4.
[2] Y. Akiyama, H. Shikagawa, N. Kanayama, T. Takarada, and M. Maeda, Chem. Eur. J. 2014, 20, 17420-17425.